Despite the approval of several vaccines to combat the COVID-19 pandemic, the disease continues unabated in many parts of the globe. New viral strains have evolved that may not be neutralized by the antibodies elicited by vaccines. Thus, there is still a need for a treatment to combat the disease.
SARS-CoV-2 is an enveloped RNA virus. Some non-structural proteins are responsible for viral replication and transcription of the viral genome. These non-structural proteins are released by two viral cysteine proteases, PLpro and Mpro (also known as 3CLpro), and these proteases have been targets for the development of antivirals.
Several inhibitors of these proteases have been investigated, but most are only slightly soluble in water. So organic solvents have to be used, which can increase their toxicity and their cost. Thus, cheap, water-soluble drugs that are readily available are needed.
Vitamin C has been known to have antiviral activity. Studies have shown it can help improve antiviral treatments. Some studies where Vitamin C was injected intravenously have shown promising results in COVID-19 treatment.
In a new study published on the bioRxiv* preprint server, researchers report the crystal structure of the complex between vitamin C and 3CLpro.
Structure of complex with protease
The team crystallized 3CLpro from the virus with ascorbate (vitamin C), and the crystals were imaged using the Advanced Photon Source at Argonne National Lab, Illinois. The structure was further refined computationally.
The structure showed an electron density difference because of the ascorbate and an additional electron density change because of trifluoroethanol, an additive in the crystallization buffer. The ascorbate electron density difference appears in the active sites near Cys-145 and His-163.
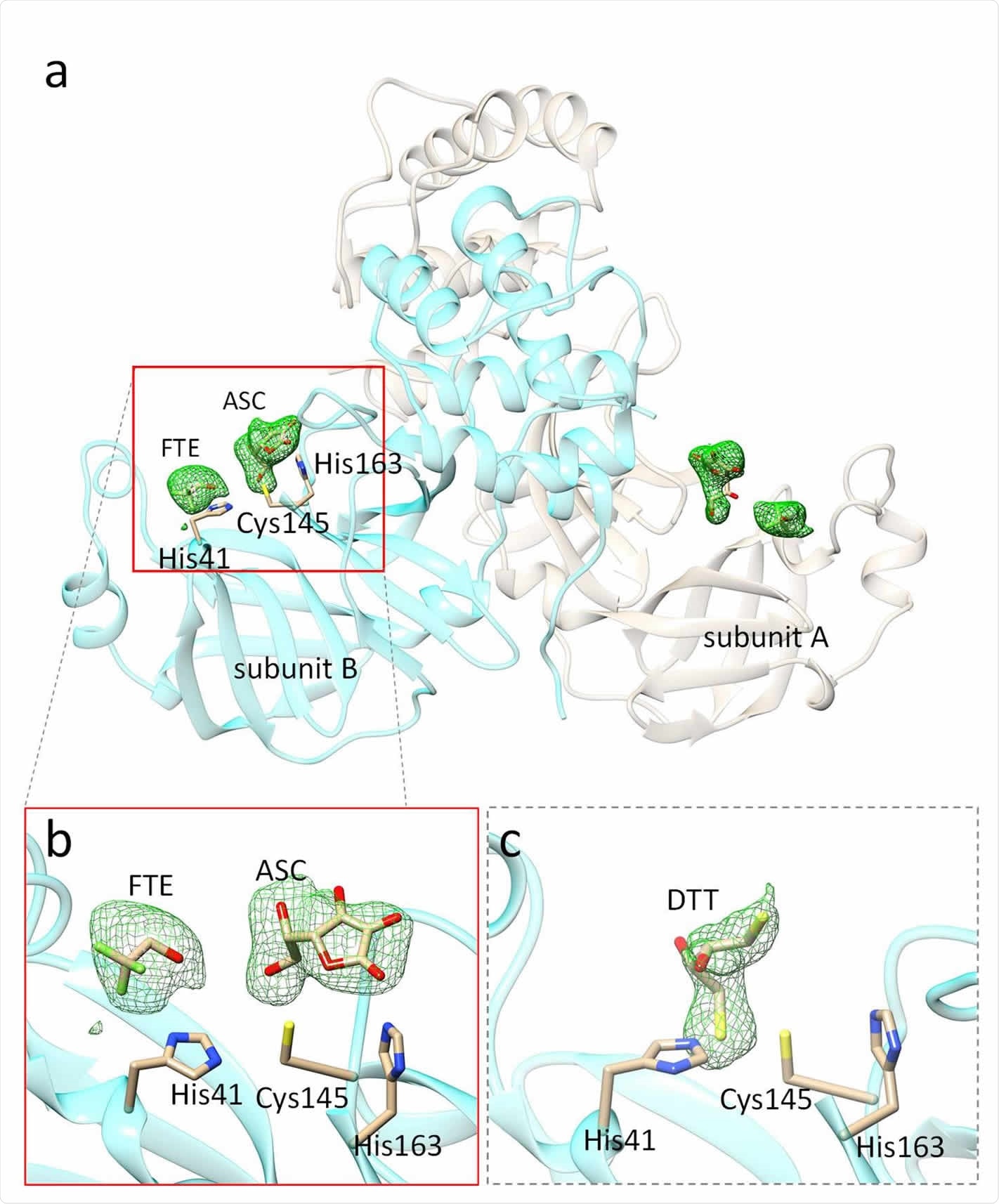
Vitamin C and DTT bound in the active site of SARS CoV-2 3CLpro. (a) The 3CLpro dimer in the asymmetric unit of the orthorhombic crystals soaked with L-ascorbate. Polder difference electron density (Liebschner et al., 2017) in the active sites is shown in green (contour: 2.5 sigma). (b) The L-ascorbate (ASC) in subunit B. The ASC interacts with the catalytic Cys-145 and is stabilized by a hydrogen bond to His-163. A trifluoroethanol (FTE) is located close to the ASC. (c) Dithiothreitol (DTT) is observed in monoclinic (C2) crystal form. It does not bind to Cys-41. It rather binds to His-41 forming a sulfenamide. Difference electron density maps shown in (a) - (c) are obtained after refining the 3CLpro without the addition of ligand. The ascorbate and DTT explain the additional electron density.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
A compound called 13b is a potent 3CLpro inhibitor. The researchers compared the position of this compound to that of ascorbate in the active site of 3CLpro.
A search in the protein data bank for ascorbate gave 40 structures that have a bound ascorbate, none of which are viral proteins. “To our knowledge, this is the first structure of a viral protein with ascorbate bound in the active site,” writes the authors. The binding of vitamin C to 3CLpro could be a possible mechanism why it could be useful for treating SARS-CoV-2 infection.
Proteases similar to the SARS-CoV-2 3CLpro can also be found in other viruses, such as picornaviruses, which cause diseases like polio and the common cold. Vitamin C could likely also help in treating other viral diseases.
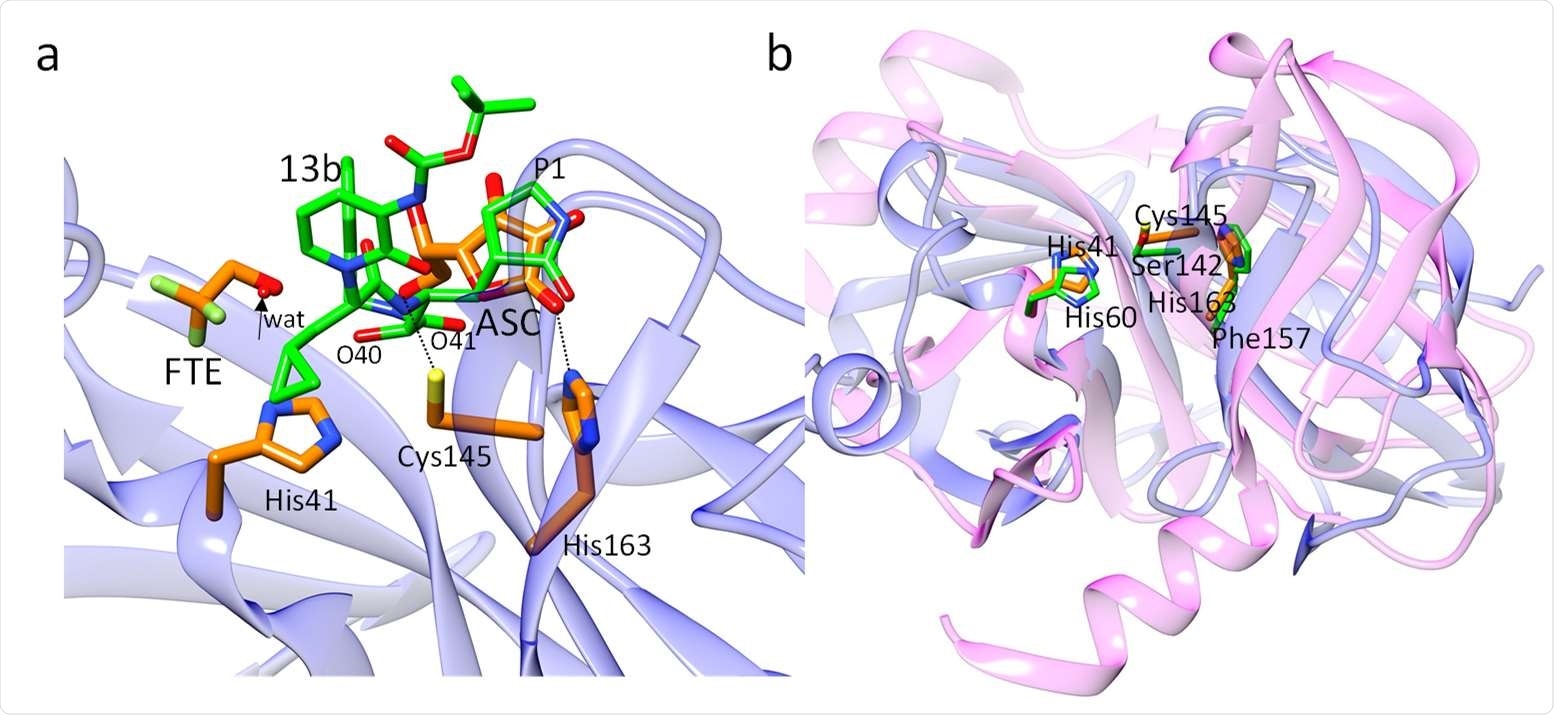
Structural comparisons. (a) Comparison of ligands in the active site of CoV-2 3CLpro. The position of the L-ascorbate molecule relative to compound 13b in 3CLpro as determined by Zhang et al., 2020. The positions of the C2’-oxygens is close to the two compound 13b oxygens O40 and O41 (marked), and the dihydroxyfuranone-ring of ASC coincides with the P1 moiety of compound 13b. The arrow points to a water molecule found in the structure of 3CLpro complexed with compound 13b. This position is occupied by the FTE oxygen. (b) Overlay of the N-terminal domain of CoV-2 3CLpro (blue ribbon, from residues 8 to 181) on the Hepatitis-C virus (HCV) serine protease (magenta ribbon). Active site residues are shown (orange for the 3CLpro, and green for the HCV protease). The active sites of both proteins are very similar. The positions of the catalytically active Cys145/Ser142 and His41/His60 are essentially identical. His163 in CoV-2 3CLpro is replaced by the structurally similar Phe157 in the HCV protease.
Vitamin C inhibits 3CLpro in vitro
The researchers also tested the activity of 3CLpro in vitro using an assay kit and by varying the ascorbate concentration. The test showed that ascorbate inhibits 3CLpro in the absence of trifluoroethanol, indicating ascorbate binding is independent of trifluoroethanol.
The half-maximal inhibitory concentration (IC50) of ascorbate is 38 mmol/L, much higher than those of other identified inhibitors. Viral load tests in cell cultures also showed ascorbate to be in the millimolar range. Studies have shown that up to 1.5 g of vitamin C per kg body mass can be given daily. It is possible that vitamin C inhibition of proteases or derivatives or direct binding to key residues such as Cys-145 could be a factor in treating COVID-19 and other viral diseases and could be a potential approach to new treatments.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.