New research led by Daniel A Keedy of City University of New York, USA, shows the structure of the main protease (Mpro) of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – the causative pathogen of coronavirus disease 2019 (COVID-19) – reacts to being heated and cooled. The temperature-dependent conformations could act as potential therapeutic targets to neutralize the virus.
The study “The temperature-dependent conformational ensemble of SARS-CoV-2 main protease (Mpro)” is available as a preprint on the bioRxiv* server, while the article undergoes peer review.
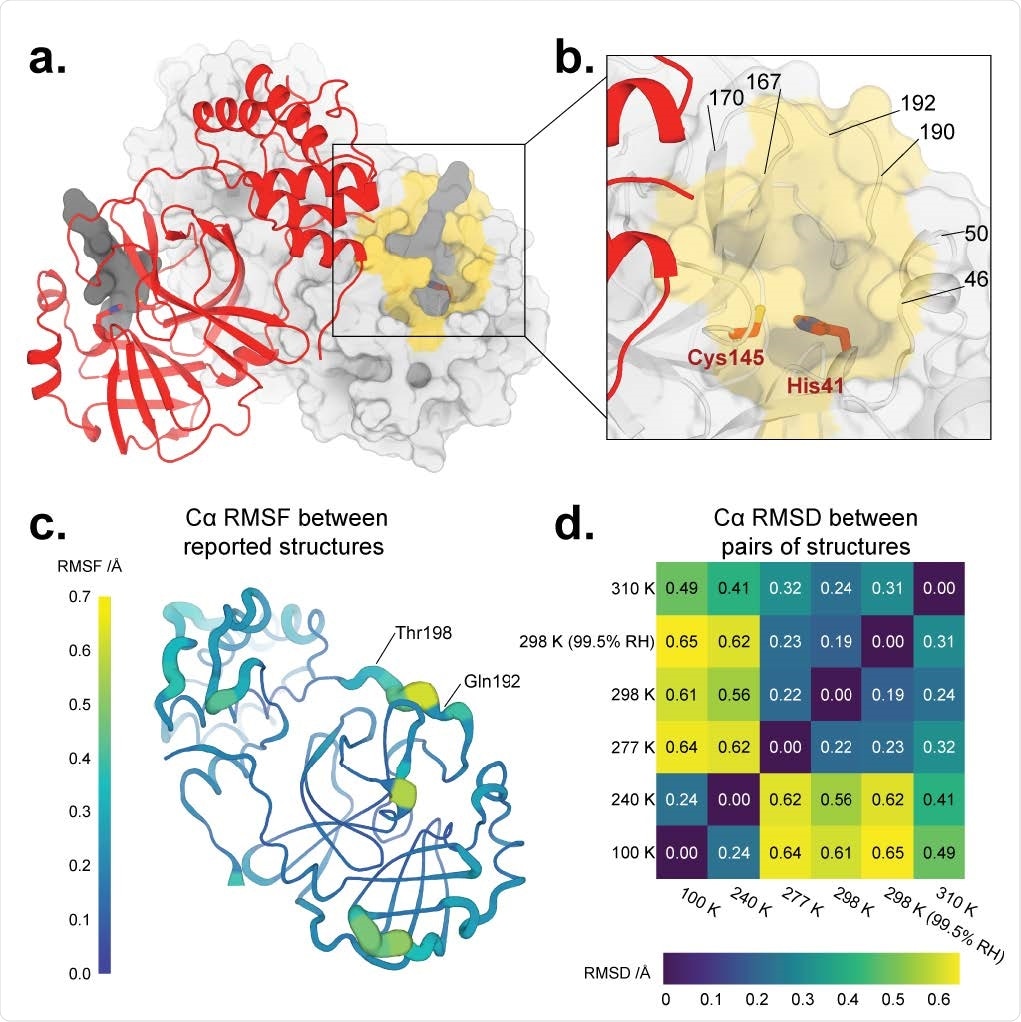
Overall structure of SARS-CoV-2 main protease at multiple temperatures. a. New X-ray crystal structure of apo Mpro at physiological temperature (310 K) (red). The biological dimer involving the other monomer (light grey surface) is constituted via crystal symmetry. The competitive inhibitor N3 from a previous structure (PDB ID: 6LU7) (semi-transparent, dark grey surface) is shown in both protomers for context. b. Close-up view of the Mpro active site region, including the catalytic dyad of Cys145 and His41 (red sticks) and highlighting residues that form the substrate-binding pocket (yellow surface). c. Cartoon putty representation of conformational variability between new Mpro structures described in this work: 100 K, 240 K, 277 K, 298 K, 298 K (99.5% RH), and 310 K. Thickness and color indicate root-mean-square fluctuations (RMSF) of Cα atom positions, from low (thin, dark blue) to high (thick, yellow). The largest differences between these structures’ backbones occur between residues 192–198. Same view as a. See also Supp. Fig. 3. d. Heatmap of pairwise Cα atom root-mean-square deviation (RMSD) between final refined structures, revealing temperature-dependent clustering (top-right vs. bottom-left).

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Temperature influences SARS-CoV-2 main protease structure
Previous research looking at SARS-CoV-2 X-ray structures has been conducted at cryogenically low temperatures or room temperature. In this study, the researchers analyzed X-ray crystal structures of the SARS-CoV-2 main protease in a range of cold and hot temperatures — 100 K, 240 K, 277 K, 298 K, and 310 K.
Results showed that temperature causes the main protease to transition when exposed to less than 240 K and more than 277 K. There were several regions that experienced conformational changes in response to the rise in temperature. This included motions of buried side chains, changes in backbone regions of the main protease, and in water molecules.
The team suggests their findings align with previous computational analyses suggesting temperature-dependent alternative conformations of the main protease.
A water molecule in the main protease active site has also been affected by temperature changes. The researchers observed the unique placement of water along the catalytic dyad, suggesting it plays some role in the catalytic process.
The water may also act as a deacylating nucleophile, suggest the researchers. The team found an acyl-enzyme intermediate mutant product-bound structure of the main protease, called 7KHP and C145A, which includes a water molecule some distance away. However, it also aligned with the collinear water molecule affected by temperature.
High humidity did not affect water on the active site of the main protease. Although, it did change the solvation shell in a nearby region.
Based on the results, the researchers suggest humidity could help as an experimental variable for crystallography and evaluating the solvent slaving’s effect on protein dynamics and solvent energetics in ligand binding.
There were also temperature-dependent conformational differences in specific main protease regions not typically observed in traditional crystallography models. The structural changes included conformational motions bridging over the active site, substrate-binding pocket, inter-domain interface, and other parts of the broad dimer interface.
The researchers suggest the conformational changes make allosteric inhibition of the main protease a potential target for antiviral therapeutics. Indeed, some areas were observed in the intramolecular network with several sites situated away from the active site but can be bound to by drugs.
Future experimental prospects
“The present study offers insights into fundamental aspects of protein structural biophysics, and may also help pave the way for new efforts toward allosteric modulation of MPRO as a strategy for COVID-19 antiviral drug design.”
The researchers propose future work on SARS-CoV-2 main protease should focus on manipulating mutations on the dimer interface to stabilize an inactive monomer, creating a new target for antiviral therapies.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.