The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has entered the second year and is marked by the emergence of several new and more infectious variant lineages. Many of these variants are also associated with immune escape.
Characterization of a rare nucleocapsid variant that affects the performance of the Quidel Sofia SARS Antigen FIA antigen 62 test
Researchers from the US recently determined the sensitivity of the Quidel Sofia SARS Antigen FIA test (Sofia 2). They found a high viral load specimen that tested negative repeatedly while using this antigen test. The study is published on the preprint server, medRxiv*.
Whole-genome sequencing of the specimen revealed two mutations - T205I and D399N - in the nucleocapsid protein of the isolate. While Sofia 2 did not detect the 6 SARS-CoV-2-positive specimens with a D399N nucleocapsid mutation available in the researchers’ lab, the Abbott BinaxNOW COVID-19 Ag Card detected all of them. However, both assays detected SARS-CoV-2-positive clinical specimens with the T205I mutation.
“We further characterized a single amino acid mutation (D399N) that affected the analytical sensitivity of the Quidel Sofia SARS Antigen FIA by approximately 1000-fold, when measured on the Sofia 2.”
On testing recombinant SARS-CoV-2 nucleocapsid with these variants, the authors found a roughly 1000-fold loss of sensitivity of the Quidel Sofia SARS Antigen FIA test for the D399N mutation. Interestingly, the BinaxNOW and Quidel Quickvue SARS Antigen tests were not affected by the mutation.
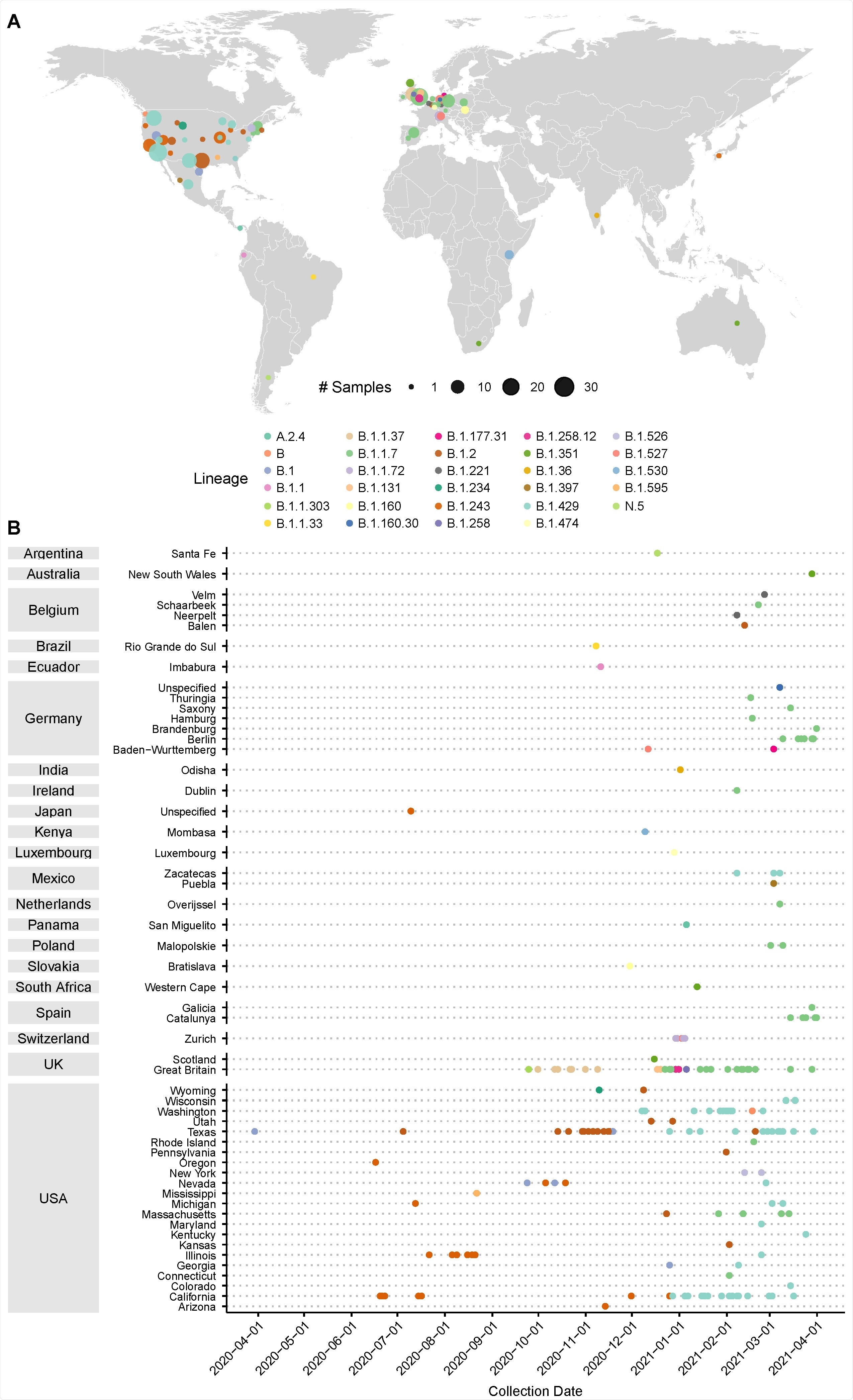
Prevalence of D399N mutations in deposited GISAID consensus sequences. A) Distribution of deposited GISAID genomes with the D399N mutation across the globe. Each dot represents sequences in a GISAID-defined subregion, with area of the dot proportional to the number of sequences. Dots are colored by PANGO lineage. B) Distribution of deposited GISAID genomes with the D399N mutation over time. Countries and subregions are indicated on the left. Each dot represents a unique deposited sequence, colored by PANGO lineage.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Rare D399N mutation affects the performance of the Quidel Sofia 2 test but does not impact the Abbott BinaxNOW test
In this study, the researchers verified the analytical sensitivity of the Quidel Sofia SARS Antigen FIA and discovered a nucleocapsid variant that considerably affected the sensitivity of this antigen test. The analytical sensitivity of the Quidel Sofia 2 (48,100 – 61,100 copies/swab) closely matches that of the Abbott BinaxNOW (40,400 – 80,600 copies/swab). However, the D399N nucleocapsid mutation affects the performance of the Quidel Sofia 2 test. Relatively uncommon, the D399N mutation is seen in only 0.02% of genomes globally.
The authors found that the single amino acid mutation - D399N - impacted the sensitivity of the Quidel Sofia SARS Antigen FIA by roughly 1000-fold. Given that there is a mutation that affects diagnostic sensitivity, the data from this study illustrates the lack of understanding of mutations and their impact on the nucleocapsid protein as compared to the spike protein of the coronaviruses.
“This mutation could further be probed by examining more specimens for which routine genome sequencing had been performed.”
Findings show the importance of routine genomic sequencing in the microbiology lab in analyzing diagnostic edge cases
The performance of antigen tests is often less well-characterized compared to molecular testing and the emergence of viral variants further effects on assay performance. Initiatives from the FDA, NIH, and CDC are increasingly trying to examine the effect of variants on the performance of diagnostic tests. However, it is worth noting that while several molecular tests have already been affected by viral variants, there are no reports of antigen tests being affected by the variants before this study over a year after authorization.
The study results show how routine pathogen genomics can be used along with clinical microbiology lab work to study diagnostic edge cases. Moreover, they highlight the importance of profiling antigenic diversity outside the spike protein in SARS-CoV-2 diagnostics.
“Although this specific story uncovered a variant that affected SARS-CoV-2 antigen testing, the ability of decentralized, widespread, routine whole-genome sequencing to uncover the genetic basis of assay performance could equally be applied to almost any diagnostic test in the clinical microbiology lab.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
A SARS-CoV-2 Nucleocapsid Variant that Affects Antigen Test Performance, Lori Bourassa, Garrett A. Perchetti, Quynh Phung, Michelle J. Lin, Margaret G. Mills, Pavitra Roychoudhury, Kimberly G. Harmon, Jonathan C. Reed, Alexander L. Greninger medRxiv 2021.05.05.21256527; doi: https://doi.org/10.1101/2021.05.05.21256527, https://www.medrxiv.org/content/10.1101/2021.05.05.21256527v1
- Peer reviewed and published scientific report.
Bourassa, Lori, Garrett A. Perchetti, Quynh Phung, Michelle J. Lin, Margaret G. Mills, Pavitra Roychoudhury, Kimberly G. Harmon, Jonathan C. Reed, and Alexander L. Greninger. 2021. “A SARS-CoV-2 Nucleocapsid Variant That Affects Antigen Test Performance.” Journal of Clinical Virology 141 (August): 104900. https://doi.org/10.1016/j.jcv.2021.104900. https://www.sciencedirect.com/science/article/pii/S1386653221001670.