The last few months have seen the emergence of new variants of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), even as other variants fade away. Some of these variants have biological or epidemiological characteristics of particular interest since they may promote the virus's rapid spread or cause more severe illness.
Earlier, three viral lineages have been classified as variants of concern (VOCs), namely the UK variant (B.1.1.7), the South African (B.1.351), and the Brazil (P.1) variant. A VOC has higher transmissibility or virulence, which potentially evades immune neutralization.
Background of B.1.620 emergence
The B.1.177 lineage spread widely in Spain first, before its transmission to the rest of Europe. Such phenomena are driven by restrictions on human movements, lack of monitoring of the outbreak, and time. In Europe, genomic sequencing projects have picked up the rapid rise to dominance of the UK strain, displacing other widely circulating strains.
In Uganda, too, the lineage called A.23.1 rose to prominence against a setting of low genomic sequencing. Such viral lineages are often observed first, not in their country of origin but as a single case or as a chain of transmission, beginning with a traveler from the host country, in another country that does sequencing on a greater scale.
As more countries launch their own SARS-CoV-2 sequencing programs, introduced strains are easier to detect since they tend to be atypical of a host country's endemic SARS-CoV-2 diversity, particularly so when introduced lineages have accumulated genetic diversity not observed previously, a phenomenon characterized by long branches in phylogenetic trees.
For instance, lineage B.1.380 was prevalent in Rwanda and Uganda for a time but was then found to give way suddenly to A.23.1. The latter was first sequenced in Uganda, which had started sequencing by then. This program then detected the first occurrences of B.1.1.7 and B.1.351.
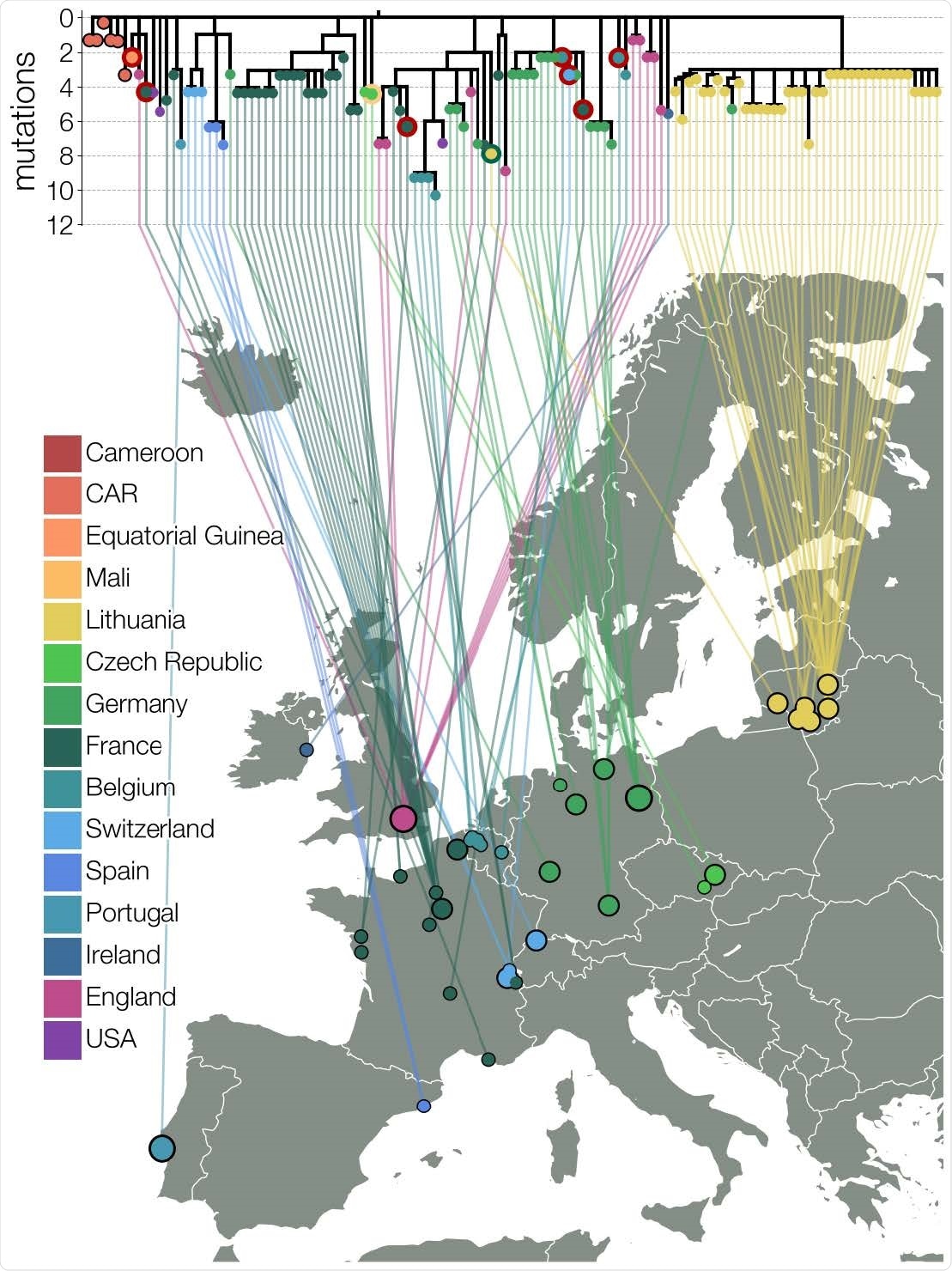
Maximum likelihood tree of lineage B.1.620 in Europe. Relationships between B.1.620 genomes, colored by country of origin (same as Figure 1) with a thicker colored outline indicating the country of origin for travel cases. At least seven genomes shown (samples collected in Belgium, Switzerland, France, and Equatorial Guinea) are from individuals who returned from Cameroon, one is from a traveler returning from Mali and one Lithuanian case returned from France. Genomes from CAR and Czechia (returning traveler from Mali) are descended from the original B.1.620 genotype, while the genome from Equatorial Guinea is already closely related to genomes found in the UK and happens to be a travel case from Cameroon. Each genome is connected to the available geographic location in Europe with the smallest circles indicating municipality-level precision, intermediate size corresponding to county-level information (centered on county capital) and largest circle sizes indicating country level information (centered on country capital).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
B.1.620 lineage in Europe
B.1.620 genomes have been found not only in Lithuania but in other places in Europe, though they do not appear to have been triggered by the same introduction event. In both Germany and France, new clades are being observed which have nearly identical genomes.
France showed a cluster of asymptomatic B.1.620 infections as part of a single chain of transmission, with another four cases outside this area. The index case is yet to be identified, but this shows community spread rather than importation.
Again, Spain and Belgium have shown the presence of B.1.620 genomes on routine surveillance sequencing.
In Lithuania, the E484K mutation in B.1.620 has been observed only in B.1.1.7 other than 13 cases of infection with B.1.351 and one with the B.1.1.318 lineage. None of these latter lineages were present in Utena county.
Many cases in Europe have had a history of travel from Cameroon, and sequencing in central African countries, namely, Central African Republic, Equatorial Guinea and the Democratic Republic of the Congo, has repeatedly thrown up B.1.620 genomes.
Study aims
In the current study, the researchers characterized the mutations of this genome. Many have already been observed in one or other of the VOCs, but not together. They also concluded that the lineage probably cropped up first in Cameroon and is likely to reach a high prevalence in Central Africa.
Lineage B.1.620 has multiple mutations
Many Cameroon genomes uploaded to the GISAID (Global Initiative for Sharing All Influenza Data) database show surprising diversity. A few share mutations with B.1.620, with the earliest to appear being, possibly, synonymous mutations at 15324 and the spike mutation T1027I. These are also present in B.1.619.
The spike E484K mutation is also found in a closely related lineage from Cameroon. All these samples were collected in January and February 2021.
The B.1.620 lineage has 23 mutations and deletions compared to the reference strain. It carries a large number of unique mutations and deletions, which are dissimilar to the closest related strain in Lithuania. Though these mutations are shared by several VOCs, this lineage does not appear to have arisen by the recombination of such strains.
The D614G mutation is now present in most circulating strains, including this one. This promotes viral infectivity, perhaps by enhancing the 'up' conformation of the spike receptor-binding domain (RBD).
Immune evasion by B.1.620
The B.1.620 lineage also has several mutations in the N-terminal domain (NTD) of the spike protein, among which some remain of unknown impact. Of the rest, all partially resist neutralization by convalescent serum and NTD-targeting monoclonal antibodies. This may point to the origin of these mutations as immune-evading viral adaptations.
The S477N and E484K mutations in the RBD are found in other VOCs, but the B.1.620 does not carry either the N501Y or K417 mutations. Both the former mutations facilitate broad evasion of antibody-mediated neutralization. They also promote high-affinity binding of the RBD to its receptor, the angiotensin-converting enzyme 2 (ACE2).
Both are on the same peripheral loop at the binding interface of these two proteins, and their presence in combination improves the energetic profile favorably relative to the reference genome.
High frequency of B.1.620 likely in Central Africa
When local sequencing programs are not robust, sequencing of infected travelers is the second surveillance option to pick up and monitor different lineages. In the current study, the researchers point out that seven B.1.620 genomes were from Cameroon-returned travelers, while six were from locally collected samples in the Central African Republic (CAR), near its Cameroon border. Thus, this lineage appears to be prevalent in this geographic region.
Adequate sequencing is being carried out in several countries neighboring Cameroon, namely, South Africa and Angola to the south, Kenya to the east, Togo, and Nigeria to the northwest. The lineage, therefore, appears to have originated within the area of central Africa bounded by the borders of these countries.
Travel histories from European cases indicate separate introductions, from Cameroon, and from Mali. In one case, at least, community spread appears to have been established, with the index patient developing the infection in Belgium well beyond the incubation period of the virus and long after his return from Cameroon. Another case had no history of travel at all.
The phylogeny of B.1.620
Phylogenetic study in Africa shows that the variant probably arose in Cameroon, and then spread to both the Central African Republic and Equatorial Guinea. It then spread to various countries in Europe through multiple introductions. It appears to have entered the USA and England, while it entered Lithuania at least twice.
Many travelers have departed from Cameroon for other African countries, indicating this lineage is likely to be widespread in Africa by now. Even with the low levels of sequencing in Equatorial Guinea and DRC, its detection supports this assumption.
What are the implications?
"The discovery of a novel lineage bearing many mutations of concern and with indications that they are introduced from locations where sequencing is not routine, is concerning, and such occurrences may become an alarming norm. The emergence of B.1.1.7 was unprecedented and has had a devastating impact on the state of the pandemic, so it is concerning that similar information gaps in global genomic surveillance still persist to this day."
Poor genomic surveillance programs are available in much of the world, while vaccines continue to be largely the preserve of developed countries. However, this disparity will plague the world for decades as the virus continues to breed new variants and creep back into previously vaccinated countries.
Control measures against the new lineages coming up are bound to be knee-jerk responses, falling short of preventing the evolution of such variants. The long phylogenetic trail of B.1.620 itself hints at gradual evolution over time, and perhaps even over vast distances, or due to selection pressures during the course of chronic infection in immunosuppressed people.
The order of mutations leading up to this lineage is also impossible to recover, making it difficult to tell if any mutations here directly promoted the occurrence of other mutations by increasing viral fitness. This is a crucial shortcoming since this lineage has multiple concerning mutations.
"Our work highlights that global inequalities, as far as infectious disease monitoring is concerned, have tangible impacts around the world and that until the SARS-CoV-2 pandemic is brought to heel everywhere, nowhere is safe for long. Additionally, we highlight the importance of collecting and sharing associated metadata with genome sequences, in particular regarding individual travel histories, as well as collection dates and locations, all of which are important to perform detailed phylogenetic and phylogeographic analysis."

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources