Viruses mutate to survive. But not all mutations are beneficial to the virus. On the other hand, some mutations such as N501Y and E484K on the spike protein propelled specific severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants to ‘variant of concern’ status, due to their ability to increase transmission and weaken the response of neutralizing antibodies.
The study authors write:
“We show that in general, transmission events between individuals likely contain genetically diverse viral particles, and we find signatures of selection governing intra-host evolution. We conclude that the analysis of shared minority variants can help to identify transmission events and give insight into the emergence of new viral variants.”
The research “Diversity and selection of SARS-CoV-2 minority variants in the early New York City outbreak” is available as a preprint on the bioRxiv* server, while the article undergoes peer review.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Genetic diversity of the virus during the early pandemic
The team collected 12 nasopharyngeal samples from 11 people who tested positive for COVID-19 infection at NYU Langone Health and NYU Grossman school of Medicine from March 6, 2020, to April 9, 2020. One individual had two samples because the collection was at two different time points. The individuals’ ages ranged from 2 weeks to 60 years old. Almost 88% of the viral load accumulated in the samples covered the SARS-CoV-2 genome.
They used variant-calling software with an allele frequency of 0.02 to identify minority variants while minimizing false positives. They mapped the 12 samples against a global tree of 10,932 global isolates and characterized genetic clades by identifying amino acid mutations prevalent among various SARS-CoV-2 strains.
The samples belonged to 2 different viral clades. About 10 samples matched to clade 20C containing the D614G, ORF1b, ORF3a, and ORF1a mutations. Identification of Clade 20C matches the dominant clade in March and April 2020 as it made up 80%-90% of viral strains.
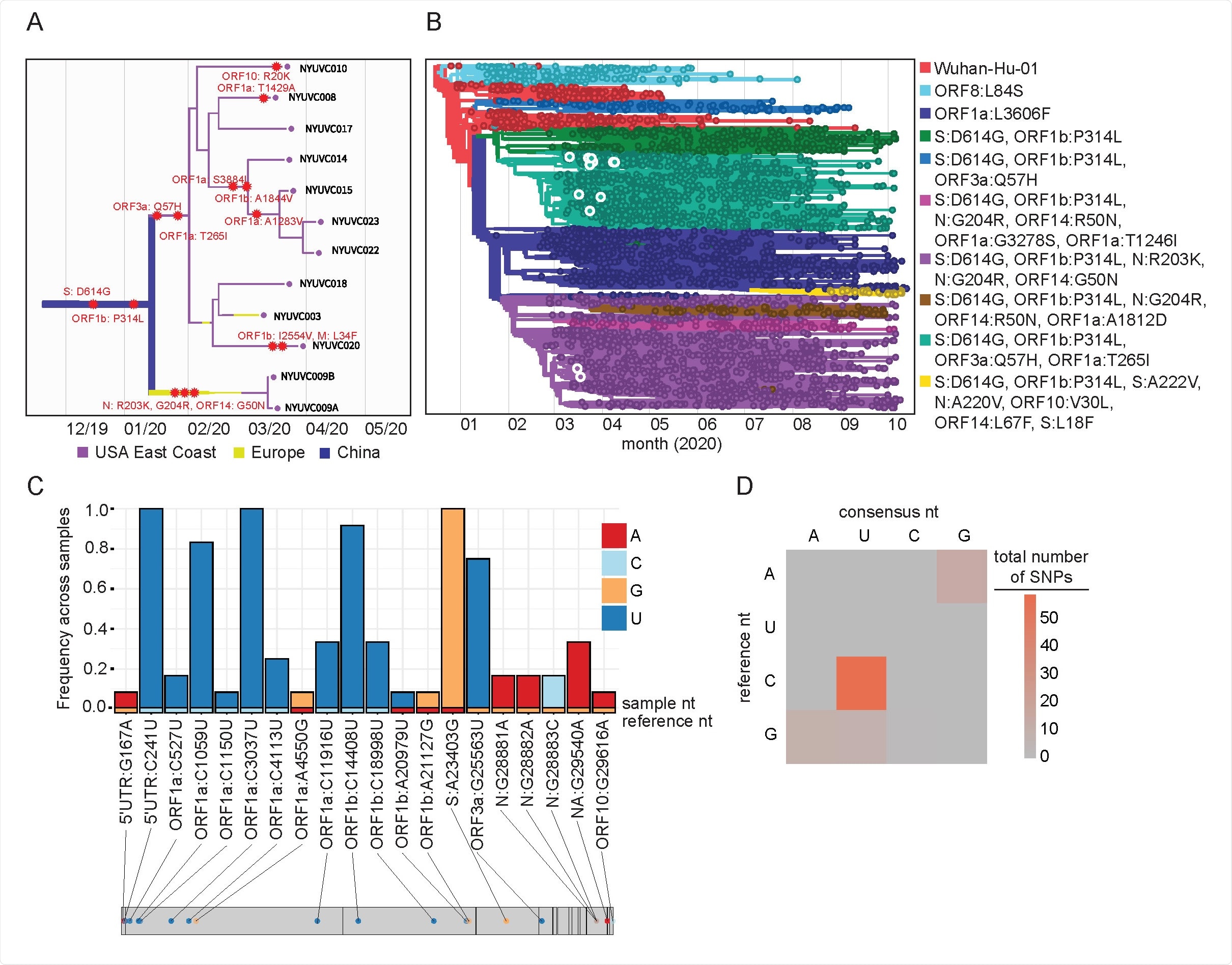
Phylogeny of New York City SARS-CoV-2 samples. (A, B) Maximum-likelihood timed strain tree reconstructed from 10932 sequences from GISAID (Methods). The tree is colored by major genetic clades, the isolates from this study are shown in detail on the left panel and highlighted in the right panel. (C) Consensus changes found with the 12 samples plotted across the SARS-CoV-2 genome. Y axis represents the frequency of a given consensus change within our cohort, where 1.0 indicates the change is found in all 12 samples. Bars are colored according to the nucleotide and the reference nucleotide (Wuhan-Hu-1) is shown along the bottom of the graph. (D) Heatmap showing the frequency of transitions and transversions represented in the identified consensus changes.
The other two sequences belonged to the same patient, and both belonged to clade 20B containing the D614G, P314L, R203K, G204R, and G50N mutations. This clade circulated about 5-10% of the time, indicating the virus was genetically diverse in the early pandemic period.
The SARS-CoV-2 ORF1a region had the most changes mutating seven different ways. Across all samples, three mutations were observed — C241U, C3037U, and A23403G. In total, there was 95 mutational change with the majority being C to U transitions.
Minority variants in New York City
The researchers found 54 minority variants across all 12 samples. About 20% of variants were found in more than one sample. The highest number of variants were in the ORF1a domain. As before, the majority of unique changes came from C to U transitions.
They also found the number of variants differed between samples, ranging from one variant in a sample to as many as 13 in another.
About 35 of the minority variants exhibited non-synonymous changes, with only one instance of a minority variant present at the same place as a consensus change in ORG1a at amino acid position 1429.
Considering most minority variants belonged to a single sample, the researchers suggest this is evidence of the randomness of errors exhibited by the viral replication process.
Minority variant features
Minority variants were common to two or more patients. Researchers were particularly interested in a pair of samples called NYU-VC-022 and NYU-VS-023 because they share several minority variants. Further analysis showed the shared variants between samples were likely caused by transmission rather than random mutation.
“NYU-VC-022 had a strongly enhanced fraction of doublet variants compared with the rest of the samples in the data set. Together, these statistics suggest a short transmission chain involving NYU-VC246 022 and NYU-VC-023 and indicate that transmission events contain a genetically diverse mix of virus particles,” wrote the researchers.
They also found the spread of minority variants in humans was possible through respiratory droplets.
Non-synonymous variants were more likely to decay faster, suggesting a loss of fitness in random and nonbeneficial mutations.
“These findings have long term implications for vaccine and drug development and set the groundwork for the exciting potential of detection of minority variants within the population before their emergence as consensus nucleotides.”

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.