The World Health Organization (WHO) has labeled the double mutant B.1.617 variant — first discovered in India —a variant of concern for its high transmission rate and ability to evade immune responses. New research posted to the preprint server bioRxiv* finds B.1.617 can modestly reduce neutralizing antibody responses generated from the Pfizer-BioNTech vaccine.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The presence of L452R and E484Q mutations on the receptor-binding domain appears to be contributing to antibody evasion. The preliminary findings show vaccinated individuals are susceptible to B.1.617 infection. Considering infections in vaccinated individuals were not severe, the international team of researchers suggests the variant of concern does not cause substantial antibody evasion — reinforcing the need for expanding vaccination to control the spread.
“At population scale, extensive vaccination will likely protect against moderate to severe disease and will reduce transmission of B.1.617 given the in vitro neutralization data we and others have presented,” concluded the researchers.
Identification of three B.1.617 lineages
The research team collected information on any whole genome SARS-CoV-2 sequences with the L452R mutation and looked for any additional mutations in the sequences.
They identified three lineages of B.1.617 with the L452R mutation. The B.1.617.1 lineage had three spike mutations on the receptor-binding domain: L452R, E484Q, or P681R. The B.1.617.2 lineage had L452R, T478K, and P681R — suggesting a loss of E484Q. E484Q reemerged in the B.1.617.3 lineage, making up 90% of the variant’s sequences.
Of all three lineages, the detection of B.1.617.1 and B.1.617.2 has steadily increased worldwide. India and 40 other countries have reported the emergence of the B.1.617 variant. The prevalence of the B.1.617.2 lineage in local and imported transmission has increased in the United Kingdom.
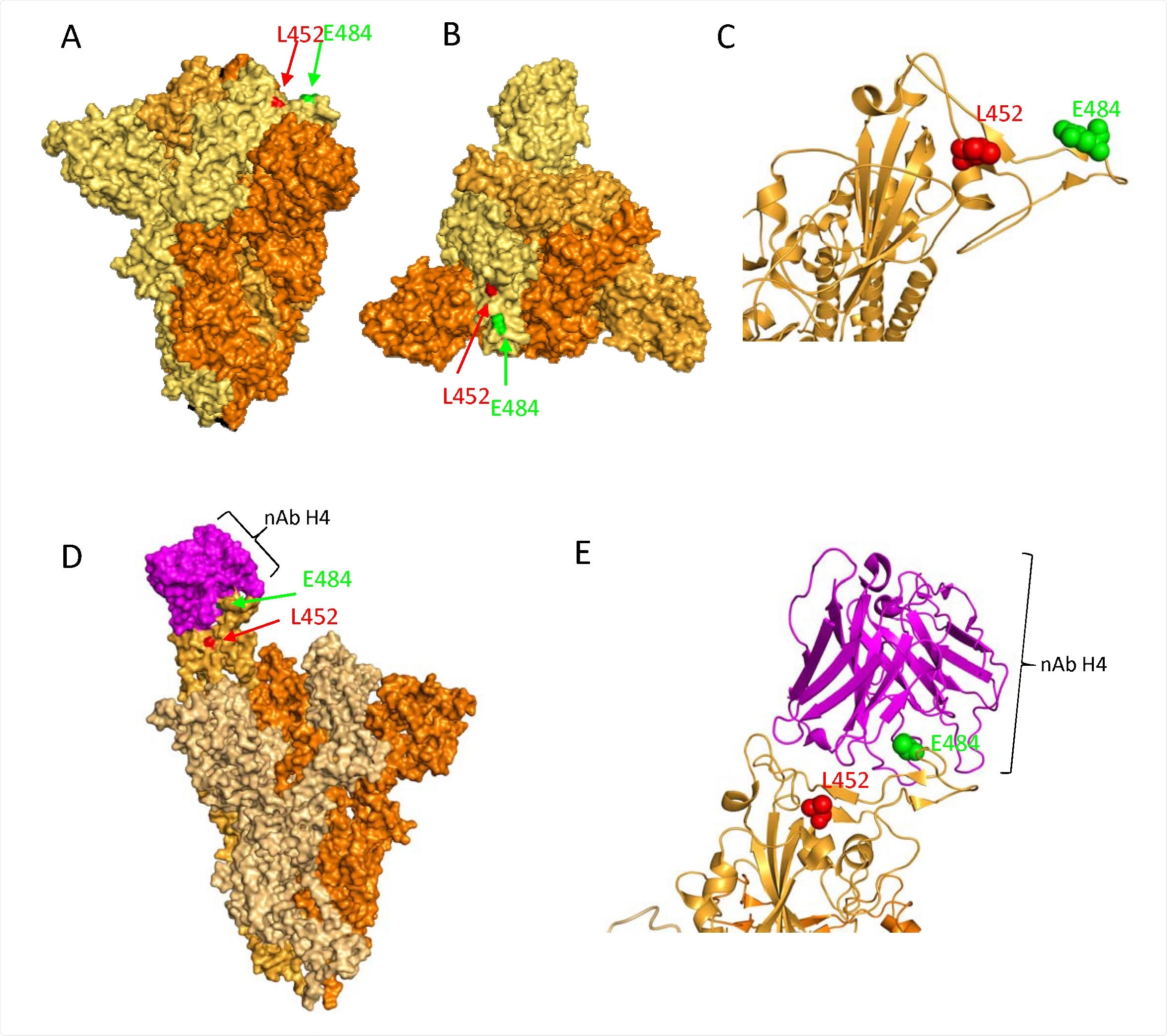
Presence of mutations L452R and E484Q in the RDB of SARS-CoV-2 spike protein. A. Surface representation of the spike protein in closed formation (PDB: 6ZGE) in a vertical view with the location of L452 and E484 mutations highlighted as red and green sphere, respectively. Each monomer in the homotrimer is coloured accordingly B. Surface representation of the same spike protein in closed confirmation in a ‘top-down’ view along the trimer axis. The residues associated with RBD substitutions L452R, and E484Q are highlighted in red and green spheres respectively, on a single monomer. C. Ribbon representation of a single monomer of the same Spike, with residues L452 and E484 highlighted as spheres coloured by element. D. Surface representation of the spike protein in open formation with neutralising antibody H4 (pink spheres, PDB: 7L58, Rapp et al, 2021) bound to one monomer of the spike protein. Residues L452 and E484 are indicated with red and green sphere, respectively. Note that E484 is partially occluded by the bound monoclonal antibody E. Ribbon representation of the interaction between the neutralising antibody H4 and the RBD of a spike monomer. Residue E484 has direct interaction with the antibody, suggesting that mutations at this site may be involved in immune escape.
B.1.617 infection in vaccinated healthcare workers
Many healthcare workers in Delhi were vaccinated with the Oxford/Astrazeneca ChAdOx1 nCoV-19 vaccine (AZD1222) in early 2021. At the time, the B.1.1.7 variant dominated the surrounding area. B.1.617 was estimated to have spread since March 2021, with infections from vaccinated individuals. A total of 33 vaccinated members — ranging from 27 to 77 years — became infected. Genomic sequencing results showed infections were derived from various SARS-CoV-2 lineages with the D614G mutation in the spike protein. However, they predominately came from the B.1.617.2 lineage.
“The dominance of B.1.617 could be explained by the prevalence of this lineage in community infection or simply reflect transmission between HCWs. The data nonetheless raise the possibility of a transmission advantage of B.1.617 in vaccinated individuals.”
There were no severe infections reported among vaccinated individuals.
B.1.617.1 variant shows partial antibody evasion
The L452R or E484Q mutations appear responsible for the moderate decrease in neutralizing antibodies produced from the Pfizer-BioNTech mRNA vaccine detecting the B.1.617.1 spike protein.
“As expected E484K conferred a ten-fold reduction in neutralization by vaccine sera, and E484Q had a slightly milder yet significant impact. When E484Q and L452R were combined, there was a statistically significant loss of sensitivity as compared to wild type, but the fold change was similar to that observed with each mutation individually with no evidence for an additive effect.”
P681R mediates viral cell entry into neighboring cells
Because the B.1.617.1 does not appear to confer high immune evasion, the researchers next looked at the variant’s infectivity rate. They infected 293T cells that had overexpression of ACE2 and TMPRSS2 — main targets of viral cell entry.
Results showed viral cell entry was similar across all mutants but lower than wild-type Wuhan SARS-CoV-2 strain.
Since the B.1.17 variant with the P681H mutation previously shown higher infectivity, they set their sights on individual mutations in B.1.617 and their effects on infectivity.
There was no significant difference in viral cell entry between L452R, E484Q/K, or the combination of the two. However, researchers did find a triple mutant with the P681R mutation increased fusion of infected cells with neighboring cells.
“We find that P681R is associated with enhanced capacity to induce cell-cell fusion and syncitia formation and that P681R alone confers this ability on the B.1.617.1 spike with RBD mutations L452R and E484Q.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Ferreira I, et al. SARS-CoV-2 B.1.617 emergence and sensitivity to vaccine-elicited antibodies. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.05.08.443253, https://www.biorxiv.org/content/10.1101/2021.05.08.443253v1
- Peer reviewed and published scientific report.
Mlcochova, Petra, Steven Kemp, Mahesh Shanker Dhar, Guido Papa, Bo Meng, Isabella A. T. M. Ferreira, Rawlings Datir, et al. 2021. “SARS-CoV-2 B.1.617.2 Delta Variant Replication and Immune Evasion.” Nature 599 (September): 1–8. https://doi.org/10.1038/s41586-021-03944-y. https://www.nature.com/articles/s41586-021-03944-y.