Researchers in the United States analyzed the non-structural protein 2 (Nsp2) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in order to better understand its function and to guide future drug discovery. The research paper is currently available on bioRxiv* preprint server.
Study: CryoEM and AI reveal a structure of SARS-CoV-2 Nsp2, a multifunctional protein involved in key host processes. Image Credit: Naeblys / Shutterstock
A causative agent of the ongoing coronavirus disease (COVID-19) pandemic, SARS-CoV-2, produces two large polyproteins after entering our cells, which are further processed into 16 individual non-structural proteins (nsp1-nsp16) by two viral proteases. They fulfill a plethora of essential viral functions - including RNA replication and double-membrane vesicle formation.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Despite their role in viral pathogenesis, many of these non-structural proteins have not yet been functionally and structurally characterized. In addition, the exact interaction interface between viral and host proteins is even less understood.
Since viruses are able to hijack central nodes in host cell pathways, obtaining molecular details of viral-host interaction may result in a better understanding of viral pathogenesis, as well as the discovery of attractive targets for antiviral treatment, since there is a lower risk of generating resistance.
The SARS-CoV-2 protein Nsp2 is involved in different viral processes; however, its exact functions remain unknown. This is why a research group, led by Dr. Meghna Gupta from the University of California in San Francisco, presented a detailed structure of the aforementioned protein in the methodologically robust new report.
Predicting protein structures
The study combines experimental data obtained by cryogenic electron microscopy with a predictive algorithm developed by Google's DeepMind, AlphaFold2, which predicts the structure of proteins.
"To our knowledge, this is the first explicit use of AlphaFold2 predictions with restraints from an experimental cryogenic electron microscopy density for model building", emphasize study authors in this bioRxiv paper.
Considering the knowledge on natural sequence variation in SARS-CoV-2 in combination with structure and mass spectrometry experiments, the researchers have successfully identified two key Nsp2 surfaces necessary for specific host interactions.
Lastly, in order to better understand which regions of the NSP2 protein are functionally significant, this research team has aligned sequences of NSP2 across beta-coronaviruses that are found in different species.
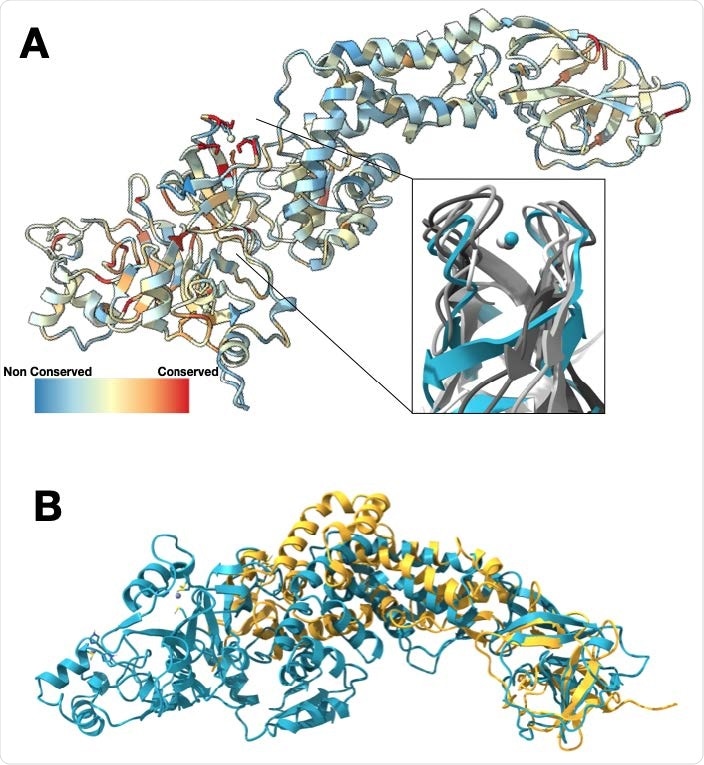
Nsp2 has a conserved zinc-binding motif but otherwise shows low conservation. (A) Nsp2 structure depicted as ribbon and colored by conservation (see methods for details). The four cysteines show the highest conservation and are indicated in red. The magnified insert shows the zinc ribbon motif of Nsp2 in cyan aligned to zinc ribbon motifs from structurally similar structures in the PDB in shades of gray (PDBs: 1JJ2, 5XON, 1QUP, 4C2M). (B) Structure of IBV Nsp2 (PDB:3LD1, yellow ribbon) aligns well to the C-terminal region of SARS-CoV-2 Nsp2 (cyan) even though it has less than 10% sequence identity.
Various roles of SARS-Cov-2 Nsp2
The resulting structure of this study shows a highly-conserved zinc ion-binding site, implying an important role for Nsp2 in RNA binding. Furthermore, detailed mapping of emerging mutations from SARS-CoV-2 variants on the resulting structure demonstrates potential host-Nsp2 interaction regions.
More specifically, this work suggests a purported role of Nsp2 in linking viral transcription that arises within the viral replication-transcription complexes to the translation initiation of the viral message.
By using structural analysis in combination with affinity-tagged purification mass spectrometry experiments, the researchers have also unveiled Nsp2 mutants that are unable to interact with different protein complexes or translation inhibitors.
Finally, different biological roles and regions of interest have been discovered on Nsp2, but mass spectrometry experiments targeting the most prevalent Nsp2 mutation (T85I) have not found any changes in host interactions of this mutant.
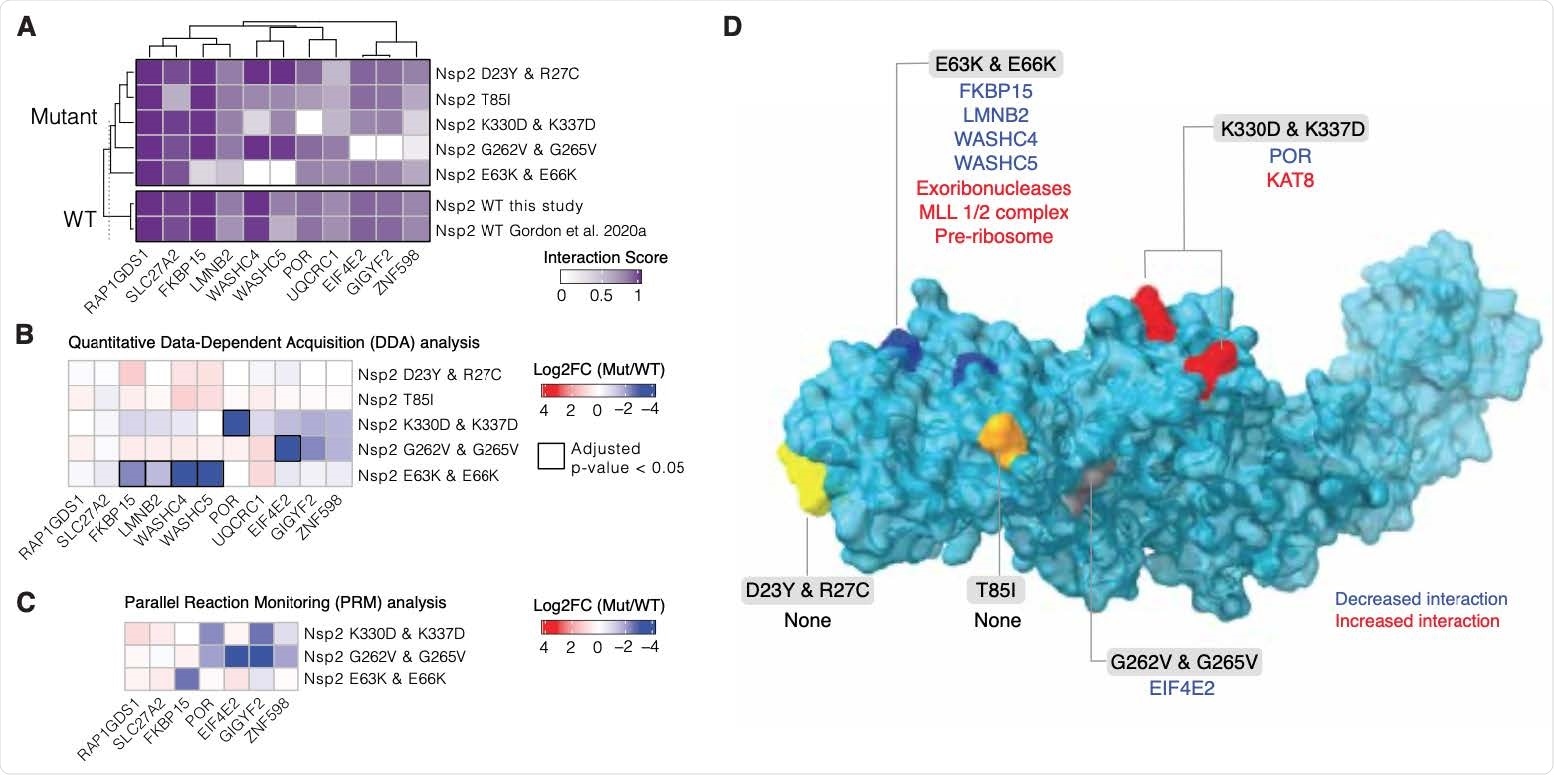
Nsp2 possesses multiple interaction surfaces for host proteins. (A) Interaction scores (average between MiST and Saint Scores) for human proteins (“preys”) deemed high-confidence interactions in at least one affinity purification (“bait”) mass spectrometry assay and detected to interact with both the wild-type Nsp2 in this study and in Gordon et al (2020a). Interaction scores range from zero to one, one being the most high-confidence. (B) Quantitative statistical analysis of data-dependent acquisition (DDA) mass spectrometry data using MSstats for interactions selected and depicted in A. Prey intensities were normalized by bait expression abundance. Log2 fold changes and BH-adjusted p-values were calculated by comparing each mutant to the wild-type from this study. Square black outlines depict adjusted p-values < 0.05. (C) Parallel reaction monitoring (PRM) analysis of select preys from B for mutants found to possess significantly-changed interactions (adjusted p-value < 0.05). (D) Nsp2 structure depicted as surface (light blue) with the mutations considered in this study depicted on the surface: E63K/E66K (dark blue), K330D/K337D (red), D23Y/R27C (yellow), T85I (orange), and G262V/G265V (grey). Lost interactions (adjusted p-value < 0.05) from data-dependent acquisition global proteomics analysis (DDA) from B depicted in blue and gained protein complexes depicted in red
Informing future drug design
The analysis of this Nsp2 structure has revealed a rapidly evolving protein surface that has potential consequences for host-virus interaction. And although global conservation among beta-coronaviruses is low, the zinc-binding motif is highly conserved.
"Collectively, the structure reported here, combined with mutant interaction mapping, provides a foundation for functional studies of this evolutionarily conserved coronavirus protein and may assist future drug design", conclude the authors of this study.
It remains to be seen how the results of this study will help in informing further research needed for the development of new drugs, but also how protein interaction mapping endeavors may reveal targets for drug repurposing.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.