Pseudouridine is the most common ribonucleic acid (RNA) modification, resulting from the isomerization of uridine (U), and is found in around 7% of all U sites except in messenger RNA (mRNA), where it presents far less frequently.
Pseudouridine levels have been found to rise in response to environmental stress, as demonstrated by mass spectroscopy and next-generation sequencing.
Quantification of the modification is difficult using these methods, though recently developed nanopore devices can be utilized to directly sequence RNA while detecting the position of epitranscriptomic modifications.
In a paper recently uploaded to the preprint server bioRxiv* by Fleming, Mathewson & Burrows (May 10th, 2021), nanopore devices are developed that allow identification of the passage of pseudouridine through a protein sensor. This further allowed the group to identify five conserved pseudouridine sites on the 3’ end of SARS-CoV-2 sub-genomic RNAs, which could potentially be beneficial to the virus in immune evasion.
Third-generation sequencing
Third-generation sequencing technologies involve passing a single DNA or RNA molecule through a pore just nanometers across and measuring the change in an electric field, allowing researchers to infer the base pair passing through. A diverse array of third-generation sequencing platforms have been developed by companies and universities around the world that operate on fundamentally different approaches.
In this case, the portable MinION device was used, which utilizes a helicase motor protein to ratchet an RNA strand from the 3’ to 5’ end through a pore. The constriction of the pore changes due to ionic and hydrophobic interactions as the strand passes through, and post-transcriptional modifications that influence these properties can be detected in this way.
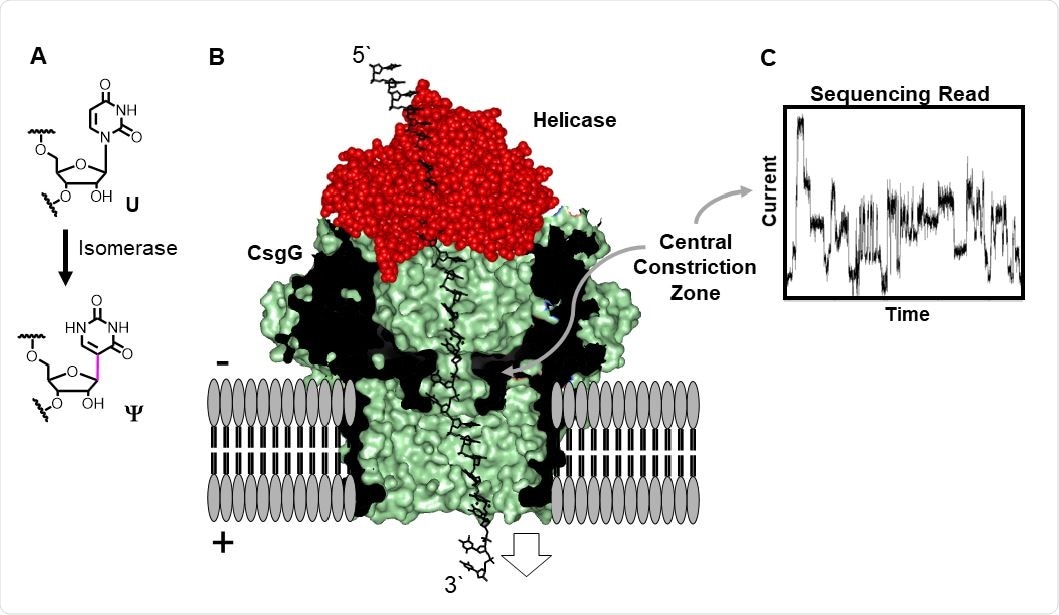
Direct RNA sequencing for Ψ by monitoring current vs. time traces in a protein nanopore-helicase platform. (A) Isomerization of U yields Ψ. (B) Structural depiction of the CsgGhelicase nanopore setup used in the R9.1.4 MinION/Flongle flowcells manufactured by ONT. (C) Example ion current vs. time trace.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The research group generated synthetic RNAs with various rates of pseudouridine modification to assess the differences in the electronic signal generated, finding that the resolution of detection for pseudouridine modifications based on changes in the electric field was around nine nucleotides, a five nucleotide window on either side of the modification.
Two helicases intended to pass the RNA stands through the pore at differing rates were utilized. It was noted that each interacted with the pseudouridine modification in differing conformations, one representing 70% of the population and the other 30%. The dwell time differences between these populations were found to more reliably evidence which conformational sub-population the pseudouridine belonged to than other detectable properties such as ionic strength, and minimizes the resolution window of modification detection down to just two nucleotides.
Similarly, unmodified U sites could be distinguished in this manner. Dwell time is also a less error-prone and situationally dependent factor than electrical signal, and the group suggests a combined approach for high-confidence analysis.
Pseudouridine bears four alterations to structure compared to uridine: a hydrogen bond donor site, introducing rigidity; a preference for a syn over an anti conformation; lesser hydrophobicity; and batter stacking with adjacent bases. The increased rigidity and improved stacking are most likely responsible for the greater dwell time observed, in addition to the differing conformation.
SARS-CoV-2 pseudouridine sites
Publically available SARS-CoV-2 RNA sequences collected from patients and bearing the pseudouridine modification were examined, including the large populations of sub-genomic RNAs generated during replication that encode for the vital structural proteins of the virus.
Five conserved sites were noted at the 3’ end of many of these short RNA sequences, potentially indicating that they are essential to replication or another stage of the virus cycle. The proteins produced by the mRNA vaccines are similarly modified with pseudouridine, and future drug discovery efforts could further exploit this structural phenomenon.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources