Researchers in the United States have developed stabilized immunogens presenting the receptor-binding domain (RBD) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that may improve the scalability of RBD-based coronavirus vaccines.
The novel SARS-CoV-2 virus is the agent responsible for the coronavirus 2019 (COVID-19) pandemic that continues to sweep the globe and has now caused more than 3.38 million deaths worldwide.
Nanoparticle immunogens presenting the RBD of the SARS-CoV-2 spike protein – the main structure that mediates host cell infection – are currently undergoing development as second-generation vaccines. They exhibit robust manufacturing features and high immunogenicity in preclinical models.
The team used previously reported deep mutational scanning (DMS) data to design variants of the RBD that showed enhanced thermal stability and resistance to aggregation, compared with wild-type RBD.
“The improvements in manufacturability, stability, and solution properties we observe could have a significant impact on the manufacturing and distribution of protein-based vaccines for SARS-CoV-2,” says the team from the University of Washington and Fred Hutchinson Cancer Research Center in Seattle.
More generally, the study also highlights the utility of comprehensive DMS data in guiding vaccine design, adds Neil King and colleagues.
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The need for robust, widely distributable vaccines
The unprecedented economic, social, and health impact of COVID-19 has demonstrated the need for reliable, robustly produced, and widely accessible vaccines against SARS-CoV-2 variations and other coronaviruses that may cause future pandemics.
There is also room to improve existing vaccines in these respects, particularly protein-based vaccines, says King and colleagues. These vaccines are yet to be widely deployed but are likely to become essential vaccines against SARS-CoV-2.
Most SARS-CoV-2 vaccines target the viral spike protein, which comprises two subunits. Subunit 1 (S1) contains the RBD that mediates host cell entry, and subunit (S2) contains the machinery that enables fusion to the cell membrane. The spike protein is considered the most important target of antibody responses to coronaviruses.
When used as a vaccine antigen, maintenance of the spike trimer in its prefusion state is vital for maximizing neutralizing antibody responses and most of the vaccines that have been authorized for emergency use utilize mutations that stabilize this conformation.
However, even with stabilizing mutations, protein-based immunogens containing the full trimeric ectodomain can suffer from instability and low expression yields, says King and colleagues. This can ultimately limit the development and scalability of protein-based immunogens.
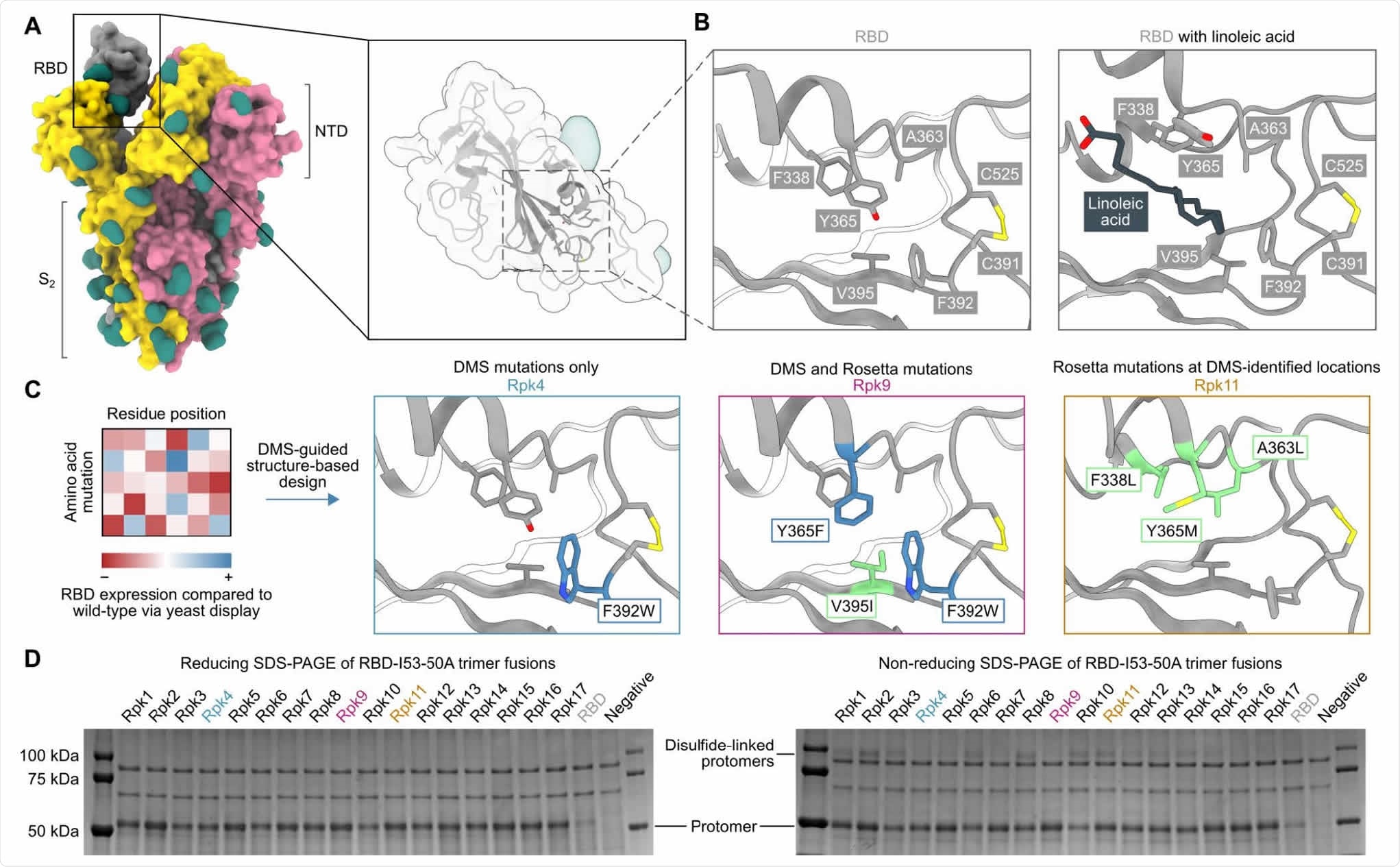
DMS-guided structure-based design of repacked (“Rpk”) SARS-CoV-2 RBDs. (A) Molecular surface representation of the SARS-CoV-2 S trimer ectodomain (PDB 6VYB), with a close-up view of the RBD (PDB 6VXX). Each protomer is colored distinctly, and N-linked glycans are rendered dark green. (B) The linoleic acid-binding pocket within the RBD, which was targeted for stabilizing mutations. The left panel shows the apo structure (PDB 6VXX) and the right panel shows conformational changes with linoleic acid (black) bound (PDB 6ZB5). (C) Mutations that increased RBD expression, identified by DMS of the RBD using yeast display (57) were used to guide Rosetta-based design of stabilized RBDs. Structural models of stabilized RBDs were generated from PDB 6VXX for Rpk4 and Rpk9, and PDB 6YZ5 for Rpk11. (D) Cropped reducing and non-reducing SDS-PAGE of supernatants from HEK293F cells after small-scale expression of stabilized RBD designs genetically fused to the I53-50A trimer. “Negative” refers to a negative control plasmid that does not encode a secreted protein.
The RBD represents a promising alternative
The RBD represents a promising alternative to the full-length spike or trimeric spike ectodomains as a target for the development of vaccines against SARS-CoV-2 and other coronaviruses.
“The SARS-CoV-2 RBD is more structurally homogeneous than metastable spike ectodomains, with far higher expression yields,” say the researchers.
Furthermore, preclinical studies have shown similar reductions in neutralizing titers against SARS-CoV-2 variants of concern for RBD nanoparticle immunogens and spike-based immunogens.
In addition, RBD antigens can be leveraged to induce neutralizing responses against the sarbecovirus subgenus and have been shown to be targeted by broadly neutralizing monoclonal antibodies.
“The potential for improved scalability and stability of RBD-based immunogens compared to spike-based immunogens, while maintaining similar immunogenicity, affirms the RBD as a bona fide antigen for use in protein-based vaccines,” writes King and the team.
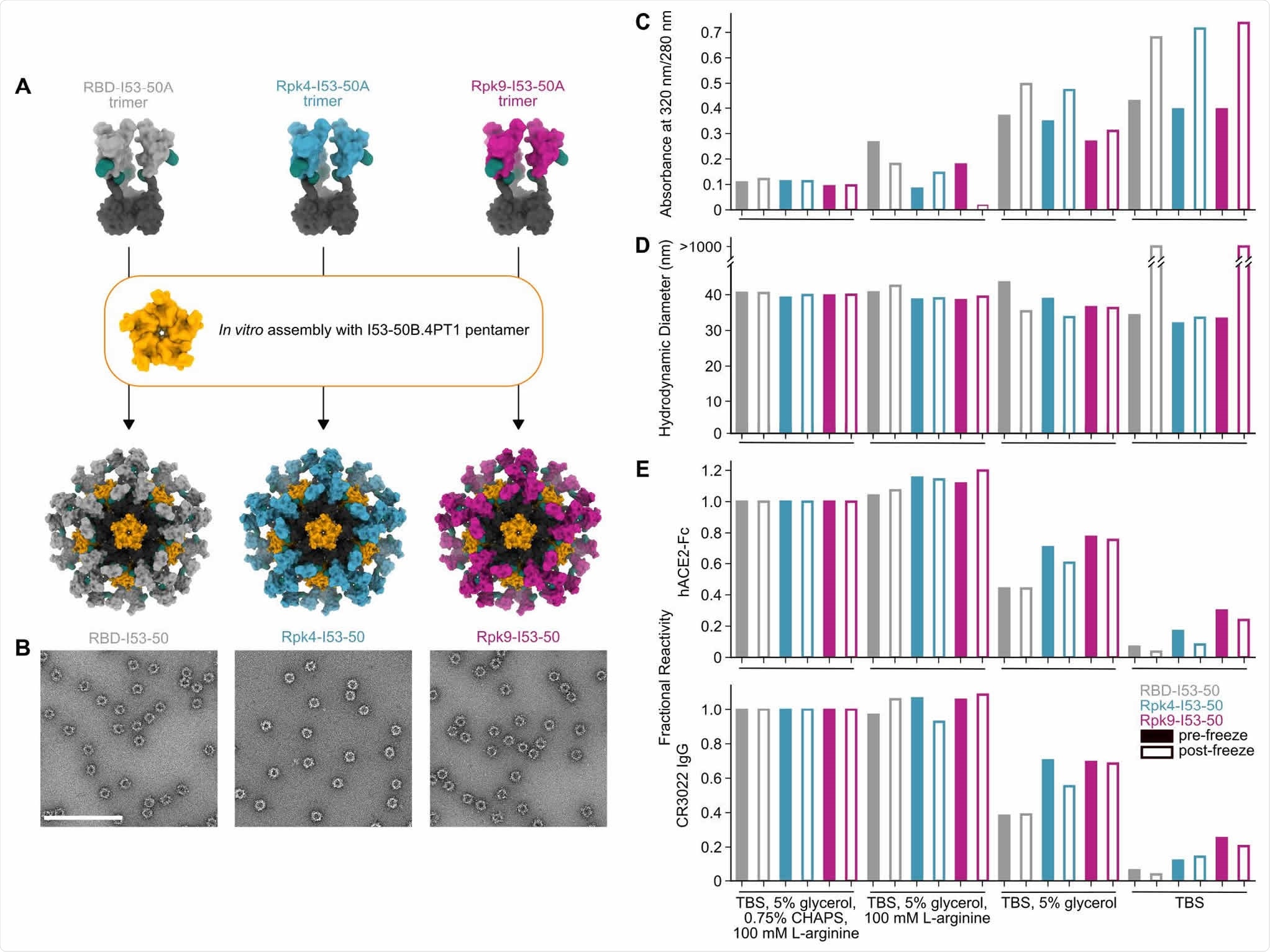
Stabilized RBDs presented on assembled I53-50 nanoparticles enhance solution stability compared to the wild-type RBD. (A) Schematic of assembly of I53-50 nanoparticle immunogens displaying RBD antigens. (B) nsEM of RBD-I53-50, Rpk4-I53-50, and Rpk9-I53-50 (scale bar, 200 nm). (C-E) show summarized quality control results for RBD-I53-50, Rpk4-I53-50, and Rpk9-I53-50 before and after a single freeze/thaw cycle in four different buffers. Complete data available in Supplementary Information 3. (C) The ratio of absorbance at 320 to 280 nm in UV-Vis spectra, an indicator of the presence of soluble aggregates. (D) DLS measurements, which monitor both proper nanoparticle assembly and formation of aggregates. (E) Fractional reactivity of I53-50 nanoparticle immunogens against immobilized hACE2-Fc receptor (top) and CR3022 (bottom). The pre-freeze and post-freeze data were separately normalized to the respective CHAPS-containing samples for each nanoparticle.
What did the researchers do?
A recent study used DMS of the SARS-CoV-2 spike RBD to identify mutations that enhance RBD expression and stability.
Now King and colleagues have combined this DMS data with a structure-based design to define stabilizing mutations in the RBD. This identified a linoleic acid-binding pocket as a source of instability.
Several stabilizing mutations to this region were found to fill this cavity and to enhance the thermal and shelf-life stabilities of RBD-containing nanoparticle immunogens. The stabilizing mutations also reduced aggregation of the immunogens while maintaining their potent immunogenicity. Corresponding immunogens displaying the wild-type RBD exhibited aggregation and loss of antigenicity.
All of the experimentally screened designs successfully improved on the expression of the wild-type RBD.
“We attribute the high success rate in part to the conservative nature of the mutations tested, but also largely to the prior validation of many of the mutations by DMS,” says the team.
“Such improvements could be crucial”
The researchers say that the thorough biochemical and biophysical characterization of RBD variants enabled them to select designs that enhanced expression, minimized off-target disulfides, improved local structural order and increased thermal, solution, and shelf-life stability, all while maintaining the potent immunogenicity of the wild-type RBD.
“Such improvements could be crucial for maximizing the scale and speed of vaccine production and buffering against unanticipated changes in the stability or solution properties of antigens derived from novel SARS-CoV-2 isolates,” they write.
Furthermore, improved resistance to denaturation and shelf-life stability at various temperatures could be particularly useful for reliable distribution in developing countries that lack cold chain infrastructures, says King and colleagues.
“Finally, the ability to reliably improve vaccine manufacturability using stabilizing mutations to the RBD may be an important tool for optimizing vaccine designs against other coronaviruses circulating in zoonotic reservoirs that threaten to cross over to humans,” concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
King N, et al. Stabilization of the SARS-CoV-2 Spike receptor-binding domain using deep mutational scanning and structure-based design. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.05.15.444222, https://www.biorxiv.org/content/10.1101/2021.05.15.444222v1
- Peer reviewed and published scientific report.
Ellis, Daniel, Natalie Brunette, Katharine H. D. Crawford, Alexandra C. Walls, Minh N. Pham, Chengbo Chen, Karla-Luise Herpoldt, et al. 2021. “Stabilization of the SARS-CoV-2 Spike Receptor-Binding Domain Using Deep Mutational Scanning and Structure-Based Design.” Frontiers in Immunology 12 (June). https://doi.org/10.3389/fimmu.2021.710263. https://www.frontiersin.org/articles/10.3389/fimmu.2021.710263/full.