Researchers in the United States have identified a shared genetic architecture between coronavirus disease 2019 (COVID-19) severity and other health conditions.
Using electronic health record data and genomic data from Veteran Affairs (VA), the Million Veteran Program (MVP), and the United Kingdom Biobank (UKBB), the team identified conditions associated with risk factors for severe COVID-19.
The study found that phenotypes associated with poor COVID-19 outcomes such as thrombotic complications also shared genetic variants associated with severe COVID-19.
Among respiratory conditions, only idiopathic pulmonary fibrosis and asthma shared genetic risk factors with severe disease, with no association observed for chronic obstructive pulmonary disease.
Anurag Verma from the Corporal Michael Crescenz VA Medical Center in Philadelphia and colleagues also found that variants associated with severe COVID-19 were associated with a reduced risk for autoimmune inflammatory conditions such as psoriasis and rheumatoid arthritis.
The team says the divergent association between severe COVID-19 and autoimmune inflammatory conditions highlights the importance of the balance between immune tolerance and immunodeficiency when considering therapeutic targets for COVID-19 therapies.
A pre-print version of the research paper is available on the medRxiv* server, while the article undergoes peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
What did the study involve?
Certain pre-existing health conditions are known to increase the risk of severe COVID-19 and death.
Genetic variants that are associated with critical illness or hospitalization due to COVID-19 have been identified in COVID-19 Host Genetics Initiative genome-wide association studies.
The team performed a phenome-wide association study (PheWAS) on 48 genetic variants associated with critical COVID-19 and 39 variants associated with COVID-19 hospitalization.
Among 455,683 MVP participants (mean age 68 years), Verma and colleagues tested genetic variants associated with severe COVID-19 for associations across 1,559 phenotypes, as well as 353,365 UK Biobank participants and 1,064 phenotypes.
Two genetic variants (rs550057, rs505922) at the ABO blood group system locus were associated with the largest number of phenotypes for health conditions, with the most significant increases in risk observed for venous embolism (27% increase) and thrombosis (31%).
“Genetic variations in ABO are an established risk factor for COVID-19 severity,” says the team. “Patients with blood group A have a higher risk of requiring mechanical ventilation and extended ICU stay compared with patients with blood group O.”
These same variations at ABO are known to be associated with several blood coagulation disorders among hospitalized COVID-19 patients, including deep vein thrombosis and pulmonary embolism.
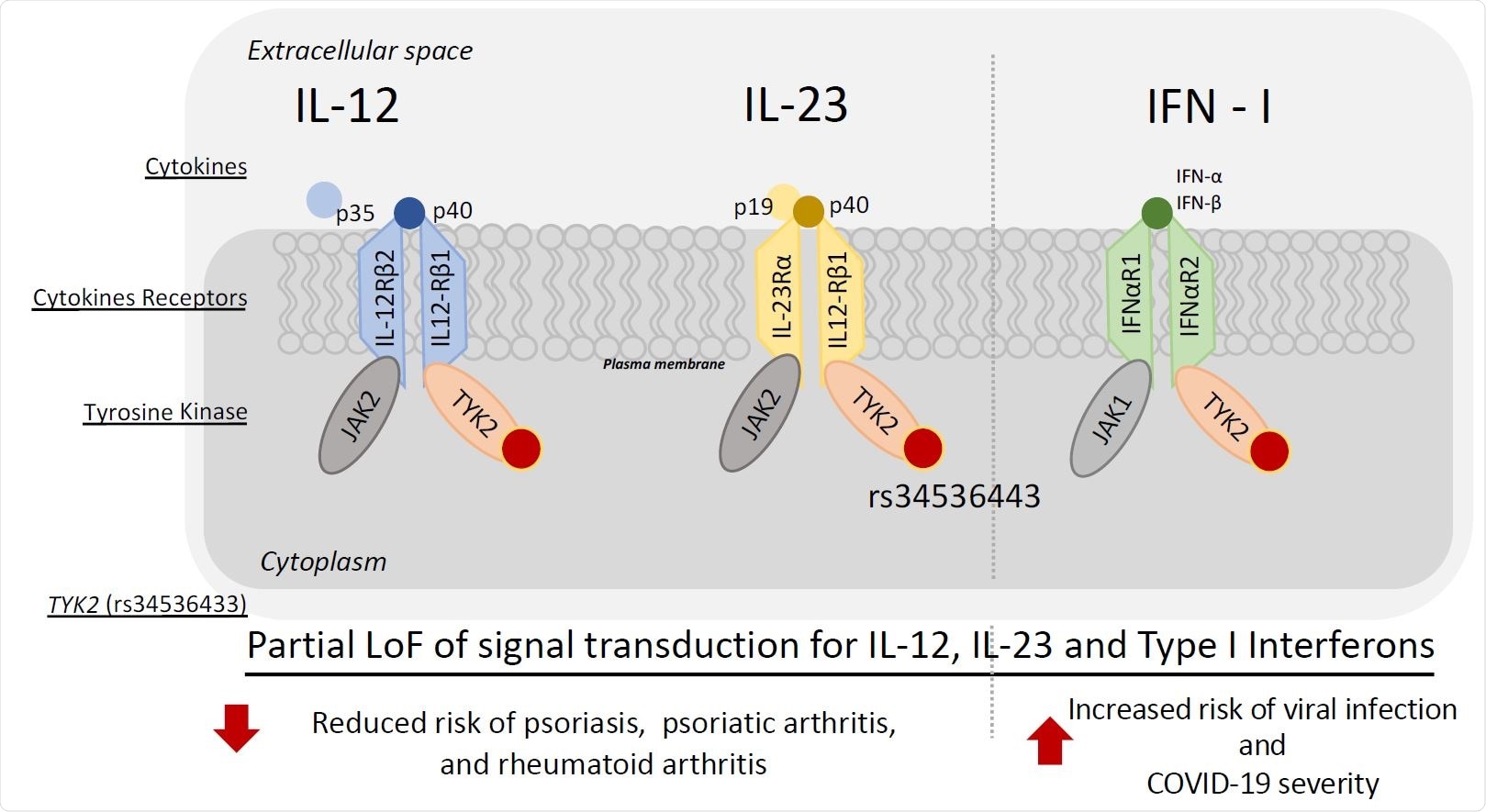
Conceptual model of the relationship between TYK2 (rs34536133) with reduced risk for psoriasis, psoriatic arthritis, rheumatoid arthritis, and simultaneous increased risk of COVID-19 severity.
What about other conditions?
Among more than 60 respiratory conditions tested, only idiopathic pulmonary fibrosis and asthma shared variants with severe COVID-19.
A single nucleotide polymorphism (SNP) rs74956615 at the ribonucleoprotein PTB-binding 1 (RAVER1) locus was associated with a 29% and 22% reduced risk for the autoimmune conditions psoriasis and rheumatoid arthritis, respectively.
The study also found that a known functional missense in tyrosine kinase 2 (Tyk2) variant (SNP rs34536443) in the region had the highest linkage disequilibrium with rs74956615, thereby suggesting that this variant drove the majority of the reduced risk.
The team says recent studies investigating potentially important pathways for COVID-19 progression have highlighted TYK2 and its downstream signaling pathway through type 1 interferons as a potential target for treatment.
“The existing literature can help explain the dual association between reduced risk of autoimmune conditions such as psoriasis and RA and increased risk of severe COVID via TYK2,” write the researchers.
The TYK2 enzyme plays a vital role in the inflammatory response and in type 1 interferon signaling – part of the innate immune response that blocks the spread of a virus from infected to uninfected cells, says the team.
What are the implications of the study?
“In this large-scale PheWAS, we identified shared genetic architecture between variants associated with severe COVID-19 and other complex conditions using data from MVP, one of the largest and most diverse biobanks in the world, with replication in UKBB,” writes Verma and colleagues.
The researchers say the divergent association between severe COVID-19 and autoimmune inflammatory conditions highlights the importance of the balance between immune tolerance and immunodeficiency.
“This balance will be important when considering therapeutic targets for COVID-19 therapies where pathways may control both inflammation and the viral host response,” they write.
“Additionally, our findings suggest that the PheWAS approach can be a useful tool to identify clinical factors related to emerging infectious diseases regarding severity or complications when genomic data are available,” says the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Verma A, et al. A Phenome-Wide Association Study of genes associated with COVID-19 severity reveals shared genetics with complex diseases in the Million Veteran Program. medRxiv, 2021. doi: https://doi.org/10.1101/2021.05.18.21257396, https://www.medrxiv.org/content/10.1101/2021.05.18.21257396v1
- Peer reviewed and published scientific report.
Verma, Anurag, Noah L. Tsao, Lauren O. Thomann, Yuk-Lam Ho, Sudha K. Iyengar, Shiuh-Wen Luoh, Rotonya Carr, et al. 2022. “A Phenome-Wide Association Study of Genes Associated with COVID-19 Severity Reveals Shared Genetics with Complex Diseases in the Million Veteran Program.” Edited by Gregory S. Barsh. PLOS Genetics 18 (4): e1010113. https://doi.org/10.1371/journal.pgen.1010113. https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1010113.