Researchers compared the nasal microbiome of coronavirus disease 2019 (COVID-19) patients, healthy individuals, and healthcare workers. These studies indicated an increase in the pathogen Pseudomonas aeruginosa present in the nasal microbiome of COVID-19 patients, which may be responsible for other secondary infections.
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spreads mainly by inhalation of airborne viral particles. The main site of initial viral replication is believed to be the nose, rather than the mouth, because of the higher expression of the receptor angiotensin-converting enzyme 2 (ACE2) in the nose.
Rapid viral replication in the upper respiratory tract can lead to infection in the lower respiratory tract and severe disease. However, many studies have shown no correlation between disease severity and viral load in the nose, suggesting that there may be other factors that contribute to disease severity.
Several studies have suggested that the microbiome in the nose and throat may play a role in viral infections. Viral infection can disrupt the microbiota and lead to coinfection, which can worsen inflammation worse and disrupt the immune response. Severe infection can also lead to a loss of both smell and taste. However, there are only a few studies that have investigated the respiratory microbiome in COVID-19 patients.
In a new study published on the bioRxiv* preprint server, researchers from the University of California Irvine investigated the effect of SARS-CoV-2 infection on the nasal microbiome.
Comparing nasal microbiomes
The team of researchers used 16S ribosomal ribonucleic acid (rRNA) gene amplicon sequencing to determine whether SARS-CoV-2 infection led to a change in the composition of the nasal microbiome. This experiment found that the nasal microbiome of COVID-19 patients was, in fact, different as compared to those who did not have the disease, as well as that which was present in the noses of healthcare workers. The nasal microbiome of COVID-19 patients and healthcare workers was richer than non-patients; however, the microbiomes of all three sample types were dominated by a select few microbes.
The nasal microbiome of a healthy person is mainly made up of Corynebacterium, Staphylococcus, Streptococcus, Dolosigranulum, and Moraxella. The authors found that many pathogenic bacteria, such as Rothia, Acinetobacter, and Pseudomonas, were common in COVID-19 patients, with particularly high levels of Pseudomonas aeruginosa reported in these patients.
Studies have shown that acute viral infections can alter the nasal microbiome and favor an increase in pathogenic bacteria. An increase in Pseudomonas has also been reported in cases of influenza, which suggests that its presence may not be specific to COVID-19 but is rather a general response to inflammatory processes.
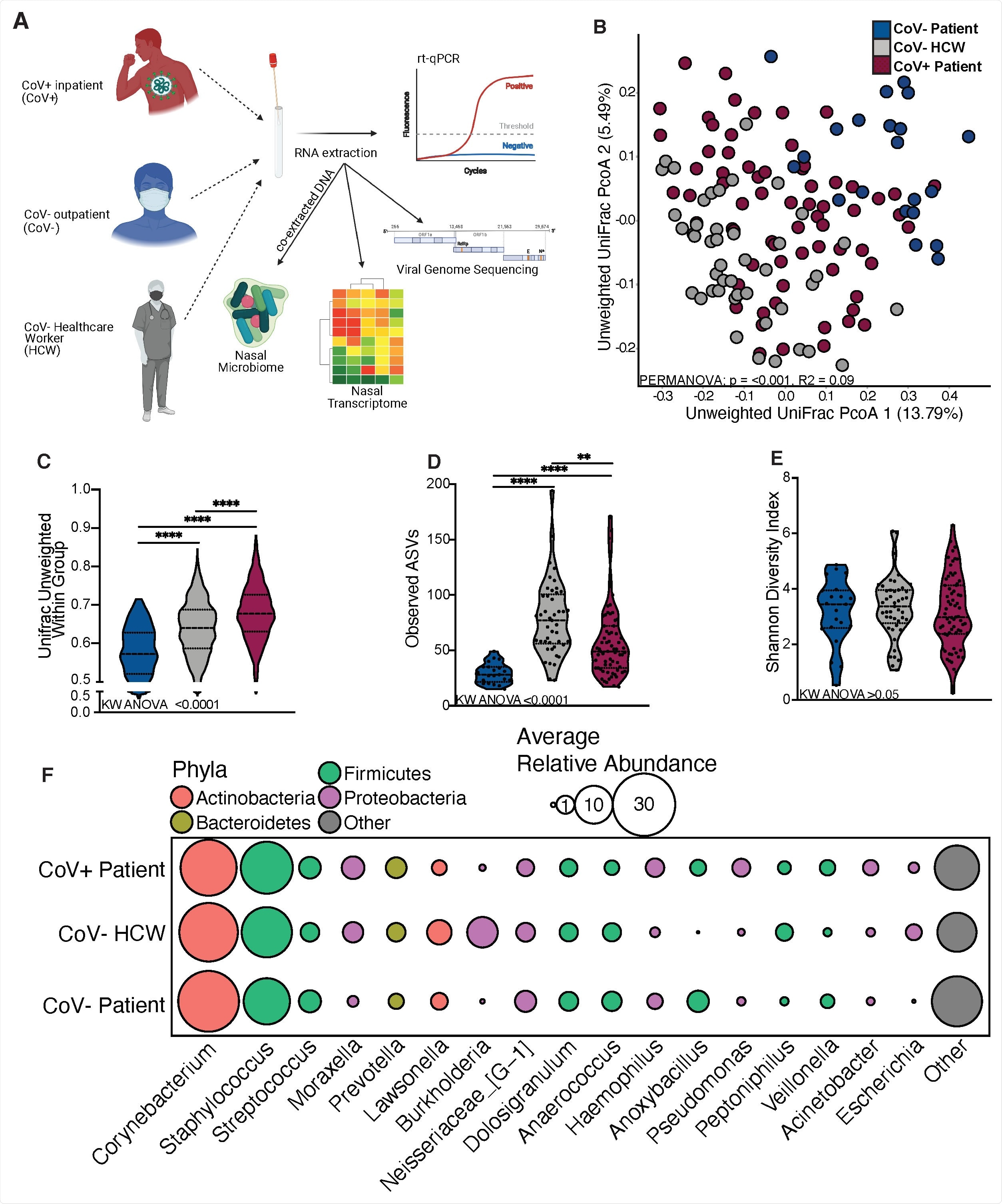
The nasal microbiome of SARS-CoV-2 infected patients is distinct. (A) Study design schematic. (B) Principal-coordinate analysis of nasal microbial communitues unweighted UniFrac distance colored by host status. The contribution of host status the total variance in the unweighted UniFrac dissimilarity matrices were measured using PERMANOVA (Adonis with 10,000 permutations). (C, D, E) Violin plot illustrating (D) average unweighted UniFrac distances (C) number of observed amplicon sequencing variants and (D) shannon diversity (E) split by host status. Significance for panels C-E was determined using Kruskal Wallis non-parametric ANOVA (p-values inset at the bottom of each panel), with Dunn’s multiple comparison * = p < 0.05, ** = p <0.01, *** = p < 0.001, **** = p < 0.0001. (F) Bubble plots of bacterial genera found at greater than 1% average abundance across the entire study population ordered from left to right in descending average abundance. The size of each circle indicates the average relative abundance for each taxa in the denoted group and the color of each circle denotes which bacterial Phyla each genera belongs to.
These results correspond to findings of secondary bacterial infections in COVID-19 patients. The nasal microbiome composition could therefore function as the primary sample type to assess a person’s risk of secondary infections following their recovery from certain viral infections.
Additionally, the California-based research group also found that viral loads in the nose did not affect the diversity of the nasal microbiome. COVID-19 patients with low viral loads were found to have a more significant amount of Streptococcus in their nasal microbiome, whereas COVID-19 patients with moderate viral loads had a greater amount of Corynebacterium. Thirdly, COVID-19 patients with high viral loads were found to have greater levels of Cutibacterium, Neisseria, and Pseudomonas species present within their nasal microbiomes.
The authors then compared the nasal transcriptomes of the test subjects and found 692 genes to be expressed differently between COVID-19 patients and healthcare workers. The upregulated genes in COVID-19-positive individuals were associated with host defense pathways. The genes that are associated with cell death, as well as those that encode for inhibitory receptors, were also upregulated in COVID-19 patients.
Comparatively, some of the genes in COVID-19 that were found to be downregulated included those that are associated with maintaining consistent internal conditions, cellular organization, and neuronal tissue processing. Furthermore, the genes that are responsible for the production of mucin in the nasal passages, as well as those that influence sensory organs, were also found to be downregulated. Taken together, these findings could explain the loss of smell and taste that has been widely reported in COVID-19 patients.
Increased pathogenic bacteria
Hospital-acquired infections remain a major challenge in the clinical setting. These infections often arise due to contaminated hospital environments and health care workers. Because of their extended exposure in hospitals, healthcare workers are common carriers of pathogenic microbes.
The authors found higher levels of pathogens like Escerichia, Klebsiella, and Burkholderia in the nasal microbiome of healthcare workers. The abundance of Acinetobacter was found to be higher in both COVID-19 patients and healthcare workers, which may be due to a possible transfer between the two.
Conclusion
The results of the present study indicate a change in the nasal microbiome of persons infected with SARS-CoV-2, with an increase in pathogens like Pseudomonas aeruginosa. One limitation of this study was the fact that only time point was assessed; therefore, future studies that compare the nasal microbiome composition of COVID-19 patients, healthy individuals, and hospital over time can improve the understanding of how the nasal microbiome changes.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Rhoades, N. S. Pinski, A., Monsibais, A. N., et al. (2021). Acute SARS-CoV-2 infection is associated with an expansion of bacteria pathogens in the nose including Pseudomonas aeruginosa. bioRxiv. doi:1101/2021.05.20.445008 https://www.biorxiv.org/content/10.1101/2021.05.20.445008v1
- Peer reviewed and published scientific report.
Rhoades, Nicholas S., Amanda N. Pinski, Alisha N. Monsibais, Allen Jankeel, Brianna M. Doratt, Isaac R. Cinco, Izabela Ibraim, and Ilhem Messaoudi. 2021. “Acute SARS-CoV-2 Infection Is Associated with an Increased Abundance of Bacterial Pathogens, Including Pseudomonas Aeruginosa in the Nose.” Cell Reports 36 (9): 109637. https://doi.org/10.1016/j.celrep.2021.109637. https://www.cell.com/cell-reports/fulltext/S2211-1247(21)01080-9.