A team of scientists from France has recently conducted a study to evaluate the sensitivity of the B.1.617 variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) to antibody-mediated neutralization. The findings reveal that compared to other circulating variants, the B.1.617 variant is resistant to therapeutic monoclonal antibodies and natural infection- or vaccine-induced antibodies. The study is currently available on the bioRxiv* preprint server.
With the progression of the coronavirus disease 2019 (COVID-19) pandemic, several new variants of SARS-CoV-2 have emerged globally. Because of acquiring multiple spike mutations, many of these variants have gained evolutionary benefits, including increased infectivity and virulence and the ability to escape host immune responses. In October 2020, a new SARS-CoV-2 lineage termed B.1.617 emerged in India, causing an exponential increase in COVID-19 cases across the country. The lineage is further divided into three subgroups: B.1.617.1, B.1.617.2, and B.1.617.3; of which the B.1.617.2 variant has been designated as Variant of Concern (VOC) because of significantly increased infectivity. However, not enough information is available on the sensitivity of this lineage to host humoral immunity.
In the current study, the scientists have investigated the sensitivity of the B.1.617.2 variant to therapeutic monoclonal antibodies, and antibodies derived from COVID-19 recovered or vaccinated individuals.
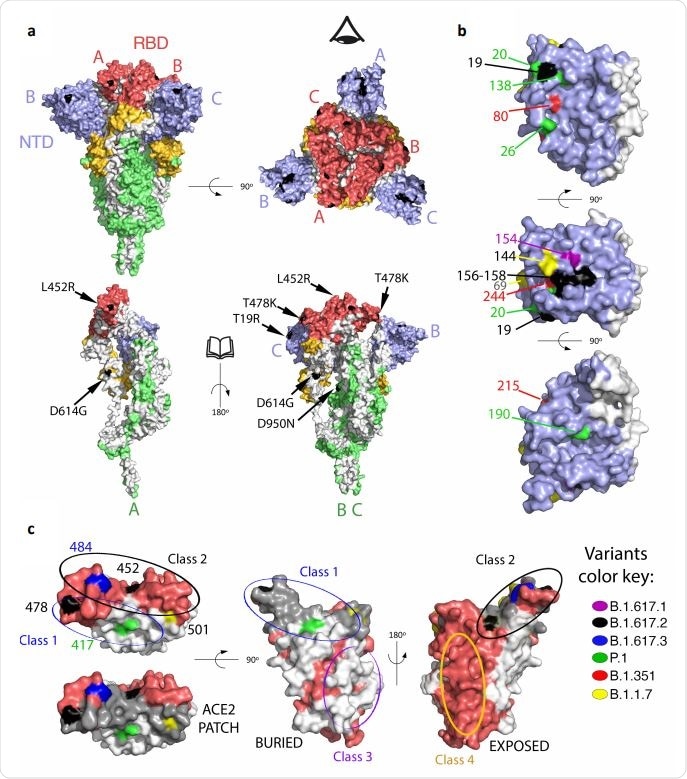
Mapping mutations on B1.617.2 and other variants of concern to the Spike surface. a. The Spike protein trimer (PDB:6XR8, corresponding to a closed spike trimer with all three RBDs in the “down” conformation) is shown with its surface colored according to domains: NTD blue, RBD red, the remainder of S1 yellow and S2 green. Interfaces between protomers were left white to help visualize the protomers’ boundaries. The three polypeptide chains in the trimer were arbitrarily defined as A, B and C, labeled to identify NTD (blue) and RBD (red) in the same protomer. Surface patches corresponding to residues mutated in the B.617.2 variant are colored in black. The top panels display two orthogonal views. The bottom panels show the trimers with subunit A in the same orientation as in the panel on top, and subunits B and C rotated 180 degrees to show the trimer interface (buried regions in the trimer are left white). The eye icon on the top right panel serves to indicate the viewpoint for the bottom right panel, after removing chain A to display internal surfaces. The mutations in B.617.2 are labeled in the bottom panel. b. Details of the NTD surface, with mutations that appeared in the indicated variants. c. RBD shown in three orthogonal views. the left panel is viewed down the surface that binds ACE2, with the surface buried in the complex with ACE2 superposed in grey in the lower panel (labeled “ACE2 patch”). The mutations on the variants are labeled. The middle panel shows the RBD surface buried in the closed spike trimer. The right panel shows its exposed surface. Note that the mutations on the RBD cluster all around the ACE2 patch. Panels were prepared with The PyMOL Molecular Graphics System, Version 2.1 Schrödinger, LLC.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Study design
In the study, the B.1.617.2 variant was isolated from the nasopharyngeal swab sample of an individual. Compared to the D614G variant of SARS-CoV-2, the B.1.617.2 variant contains nine spike mutations, including five mutations in the N-terminal domain (NTD), 2 mutations in the receptor-binding domain (RBD), 1 mutation in the furin-cleavage site, and one mutation in the spike S2 subunit.
To examine the immune escape potential of the variant, four clinically approved anti-RBD antibodies along with four other anti-RBD antibodies and four anti-NTD antibodies were included.
For antibody isolation, serum samples were collected from 56 COVID-19 recovered individuals and 28 vaccinated individuals. Of vaccinated individuals, 16 received the mRNA-based Pfizer vaccine and 12 received the adenovirus-based AstraZeneca vaccine.
Genetic characterization of B.1.617.2 variant
The spike mutations identified in the B.1.617.2 variant showed a similar distribution pattern as other VOCs. Besides the D614G mutation, the D950N mutation was identified in the trimer interface. The majority of NTD mutations were present in the “supersite,” which is the primary target of potent anti-NTD neutralizing antibodies. Similarly, all RBD mutations were identified in the periphery of the angiotensin-converting enzyme 2 (ACE2) binding surface, indicating that the B.1.617.2 variant acquires these mutations to avoid antibody binding without hampering ACE2 binding.
Monoclonal antibody-mediated neutralization of B.1.617.2 variant
All tested therapeutic antibodies (Bamlanivimab, Etesevimab, Casirivimab, and Imdevimab) showed high potency in neutralizing the D614G variant. Except for Etesevimab, the other three antibodies actively neutralized the B.1.1.7 variant as well.
Regarding B.1.617.2 variant, all antibodies except Bamlanivimab retained their active virus neutralization potency. The acquisition of L452R mutation could be the escape mutation for Bamlanivimab.
Of four other anti-RBD antibodies, three efficiently neutralized the B.1.617.2 variant. In contrast, three out of four anti-NTD antibodies failed to neutralize the B.1.617.2 variant, with the fourth one showing modest neutralizing efficiency.
The flow cytometric analysis to determine antibody–variant interaction revealed that the B.1.617.2 variant developed resistance to antibody-mediated neutralization primarily by preventing antibody–epitope binding.
Convalescent sera-mediated neutralization of B.1.617.2 variant
The serum samples obtained from COVID-19 recovered individuals after six months of symptom onset were tested against the B.1.617.2, D614G, B.1.1.7, and B.1.351 variants. Compared to D614G and B.1.1.7 variants, the B.1.617.2 variant showed 4 – 6 times less sensitivity to neutralization by convalescent sera.
Similarly, serum samples obtained from 48 convalescent healthcare workers after 12 months of symptom onset showed a six-fold lower potency in neutralizing the B.1.617.2 variant compared to the B.1.1.7 variant. Among healthcare workers, 13 received the 1st dose of AstraZeneca, Pfizer, or Moderna COVID-19 vaccine. The sera obtained from vaccinated healthcare workers showed a 130-fold increase in neutralizing antibody levels against the B.1.617.2 and B.1.1.7 variants. These findings reveal that the B.1.617.2 variant is comparatively less sensitive to neutralization by convalescent sera obtained 1 year after infection.
Vaccination-mediated neutralization of B.1.617.2 variant
The sera obtained from individuals completely immunized with the Pfizer COVID-19 vaccine showed 3-times and 16-timer lower potency in neutralizing the B.1.617.2 and B.1.351 variants, respectively, as compared to the B.1.1.7 variant. In contrast, the sera obtained from individuals immunized with a single dose of the AstraZeneca COVID-19 vaccine showed significantly lower potency in neutralizing the B.1.617.2 variant.
Study significance
The study findings reveal that the recently emerged B.1.617.2 variant of SARS-CoV-2 is partially resistant to neutralization by anti-RBD and anti-NTD monoclonal antibodies and polyclonal antibodies induced by natural SARS-CoV-2 infection or vaccination.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources