While effective vaccines against coronavirus disease (COVID-19) are being administered globally at an unprecedented pace, there is still a dire need for antiviral therapies, especially considering the emergence of antigenically distinct SARS-CoV-2 variants of concern across the world.
Antivirals can be grouped into two basic categories, host-targeting and virus-targeting, both of which warrant a thorough understanding of the molecular mechanisms used by viral agents to replicate in cells.
Furthermore, we are aware that coronaviruses replicate by using a well-established series of molecular events, where host factors are present at every step – representing in turn potentially druggable targets.
Taking into account the need for an adequate understanding of genome-wide screens in the identification of host factors necessary for viral replication, as well as the tremendous global impact of the current COVID-19 pandemic, it is of no wonder that many research groups use this approach to study SARS-CoV-2.
Consequently, a research group from the US – led by Dr. Marco Grodzki from the Department of Molecular Genetics and Microbiology, College of Medicine at the University of Florida – performed a global analysis of interactions between the host and human coronaviruses from genome-wide screens in two different cell lines.
Knockout screens and inhibitor testing
More specifically, this research group pursued genome-scale CRISPR knockout screens in two cell lines with SARS-CoV-2 and human coronavirus OC43 that causes the common cold. The cell lines they have used were Vero E6 (kidney epithelial cells from an African green monkey) and HEK293T (human embryonic kidney cells) ectopically expressing ACE2 receptor.
They have also determined whether specific pathways and gene products identified in these screens could be targeted with commercially available inhibitors to halt the infection with human coronaviruses. Many genes involved in cell cycle regulation were tested, and viral genome copies were quantified in cells treated with each compound.
Finally, the researchers have also performed a comprehensive comparative analysis of all published genome-wide SARS-CoV-2 screens to date in an effort to identify salient targets and inform further drug development.
Antivirals targeting recognized host factors
In short, genome-wide CRISPR screens in Vero E6 and HEK293T-hACE2 cells identified specific host factors required for the infection with human coronaviruses. A subset of gene candidates that promote viral replication has also been validated.
Moreover, drugs targeting selected host factors displayed in vitro efficacy against SARS-CoV-2, which included various cell cycle inhibitors, one endocytosis inhibitor and one TANK-binding kinase 1 inhibitor. These drugs could be repurposed for the treatment of COVID-19 in the near future.
Host factors involved in cell cycle regulation were enriched in the screens that were pursued in this study, as were some other key components of the programmed messenger RNA decay pathway. In accordance with the obtained results, cell-type-specific differences in the use of lysosomes for human coronavirus egress have also been suggested.
Towards the understanding of key interactions
“Our studies substantiate and expand the growing body of literature focused on understanding key human coronavirus-host cell interactions”, say the authors of this bioRxiv paper.
“The fairly limited redundancy in proviral factors identified across our study and other published studies using genome-wide CRISPR screens highlights the extensive scope of these interactions and suggests that even more host factors remain to be discovered”, they add.
In any case, differences in cell types and variable infection conditions unquestionably influence the outcomes of screens; hence, this could be a way to obtain new insights into subtle mechanisms and characteristics of SARS-CoV-2 replication.
Therefore, meticulous molecular studies that test hypotheses akin to this one (and stemming from kindred genome-scale CRISPR screens) represent a pivotal next step. Additional work is also needed to determine the mechanism of action of novel compounds before they can be used in the clinical milieu.
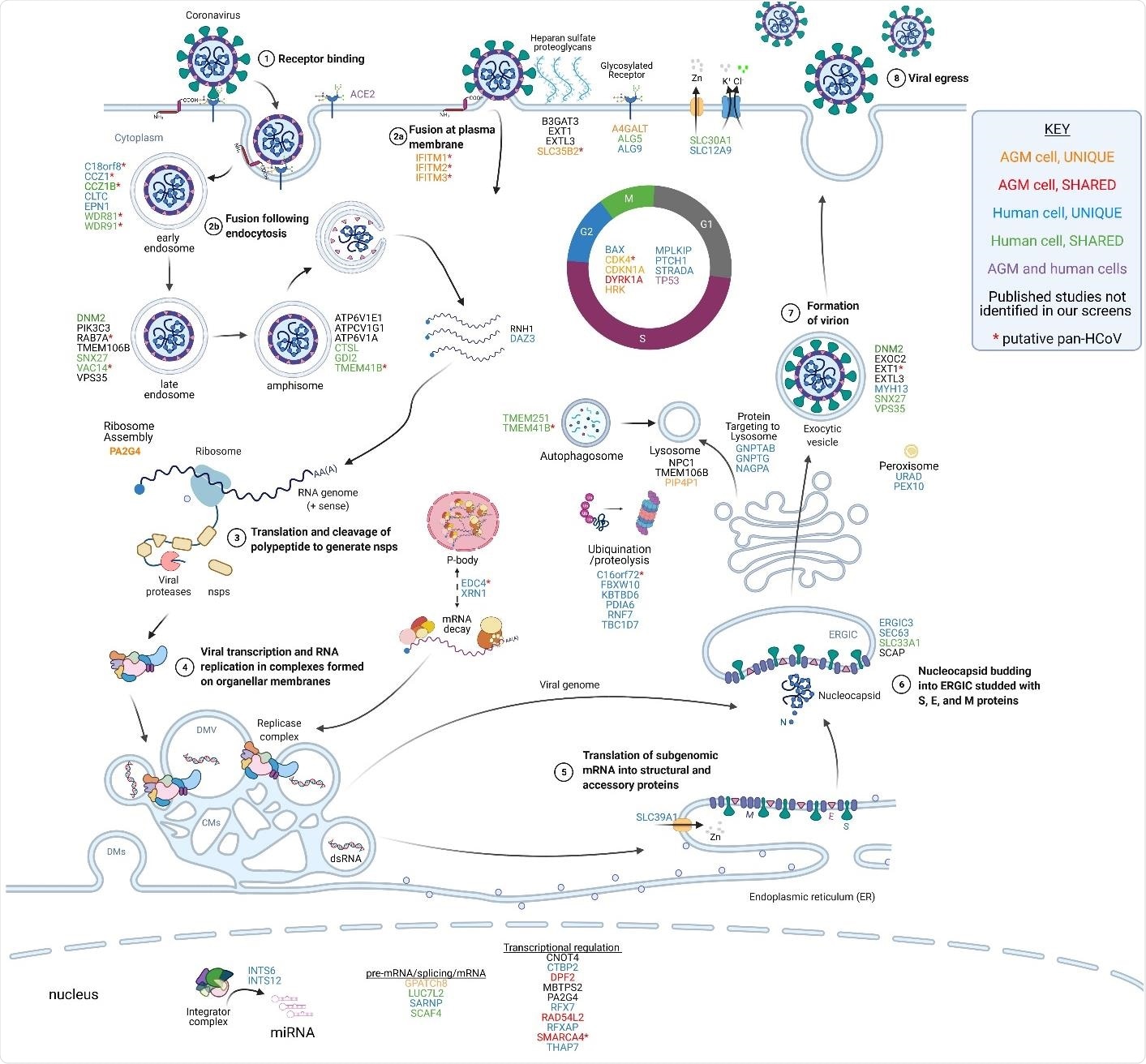
Summary of genes found in this and other studies and their potential roles in the SARS-CoV-2 life cycle. The host factors identified in CRISPR screens are presented adjacent to the putative stage of viral replication where they function. The genes are color-coded based on their identification in our and other published studies, as indicated in the legend. Candidate pan HCoV host factors are indicated with red asterisks. The virus replicates through a series of well defined molecular steps. 1-2) After virion binding to ACE2, SARS-CoV-2 can fuse at the plasma membrane or following endocytosis. Heparan sulfate proteoglycans enhance viral attachment to cells so host factors involved in heparan sulfate biosynthesis (B3GAT3, EXT1, EXTL3, SLC35B2) and glycosylation (A4GALT, ALG5, ALG9) may play a role in viral entry. The IFITM proteins are proposed to promote fusion at the cell surface but inhibit fusion in endosomes. Host factors involved in endocytosis (C18orf8, CCZ1, CCZ1B, CLTC, EPN1, WDR81, WDR91), vesicular transport (DNM2, PIK3C3, RAB7A, TMEM106B, SNX27, VAC14, VPS35), and amphisome maturation/lysosome fusion (ATP6VIE1, ATPCV1G1, ATP6V1A, CTSL, GDI2, TMEM41B) likely facilitate virion uncoating. 3) The positive-sense RNA genome is then translated to produce the nonstructural polyproteins which are co-translationally cleaved to form the mature nsps. Certain host factors like RNH1 and DAZ3 may serve to protect the viral genome from degradation by host enzymes. 4) The nsps form the viral replicase which assembles on organellar membranes to form the replication and transcription complexes (RTCs) where progeny genomes and structural/accessory protein transcripts are produced, respectively. P-body components EDC4 and XRN1, identified in this study, may play a role in the maintaining viral RNA stability or assembly of the RTC. 5) Structural and accessory proteins are translated, and structural proteins insert into the ER membrane. ER-localized SLC39A1 may play a role in this process. 6) Nucleocapsids bud into the ERGIC, potentially aided by host factors ERGIC3, SEC63, SLC33A1, and SCAP. 7) Progeny virions form as they traverse through the Golgi and structural proteins are glycosylated. 8) Virions exit the cell through either typical exocytosis (DNM2, EXOC2, EXT1, EXTL3, MYH13, SNX27, VPS35) or nonclassical lysosomal egress (GNPTAB, GNPTG, NAGPA, NPC1, TMEM106B, PIP4P1). Numerous host factors with less obvious direct roles in promoting steps in the viral life cycle have also been identified in CRISPR screens. For example, numerous factors regulating the cell cycle (BAX, CDK4, CDKN1A, DYRK1A, HRK, MPLKIP, PTCH1, STRADA, TP53) were identified in our screens in AGM and human cells. Furthermore, multiple nuclear-localized host factors including diverse transcriptional regulators and two components of the integrator complex (INTS6, INTS12) were identified. Overall, the large number of diverse host factors that promote SARS-CoV-2 replication illustrates the large-scale exploitation of cellular processes required for successful viral propagation. Adapted from BioRender template titled Life Cycle of Coronavirus generated by the Britt Glaunsinger laboratory.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Grodzki, M. et al. (2021). Genome-scale CRISPR Screens Identify Host Factors that Promote Human Coronavirus Infection. bioRxiv. https://doi.org/10.1101/2021.06.04.447090, https://www.biorxiv.org/content/10.1101/2021.06.04.447090v1
- Peer reviewed and published scientific report.
Grodzki, Marco, Andrew P. Bluhm, Moritz Schaefer, Abderrahmane Tagmount, Max Russo, Amin Sobh, Roya Rafiee, Chris D. Vulpe, Stephanie M. Karst, and Michael H. Norris. 2022. “Genome-Scale CRISPR Screens Identify Host Factors That Promote Human Coronavirus Infection.” Genome Medicine 14 (1). https://doi.org/10.1186/s13073-022-01013-1. https://genomemedicine.biomedcentral.com/articles/10.1186/s13073-022-01013-1.