Researchers have developed a rapid assay to rapidly detect neutralizing antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) using an imaging technique they developed for the Ebola virus.
In addition to respiratory disease, SARS-CoV-2 can result in cardiovascular disease, severe respiratory illnesses, and neurological disorders. Some patients also have long-term symptoms from the infection.
During severe infection, it seems that the type of antibody produced switches quickly from Immunoglobulin M (IgM) to Immunoglobulin G (IgG) and Immunoglobulin A (IgA), with a slow switch in the type associated with patients needing hospitalization. The type of antibody is important in controlling the disease, as is the virus protein targeted by the antibodies. Strong antibody response to the SARS-CoV-2 nucleoprotein is associated with severe disease.
Rapid detection and characterization of SARS-CoV-2 antibodies are needed because antibodies are an important indicator of the degree of protection of the patient. Additionally, they can be used to study vaccines and develop monoclonal antibody therapies against COVID-19.
In a study recently published in the journal Viruses, researchers from the National Institutes of Health in the United States and A. T. Kearney Inc. report a SARS-CoV-2 neutralization assay that is semi-throughput, uses an imaging system and can be used for different virus variants.
Adapting the fluorescence assay for SARS-CoV-2
The researchers adapted a fluorescence reduction neutralization assay (FRNA), previously developed for measuring neutralizing antibodies to the Ebola virus and the MERS-CoV, for use with SARS-CoV-2. The FRNA method uses only a small sample volume in a 96-well plate. This also makes it easy to automate. The output fluorescence is measured using an optical reader.
Unlike other assays for neutralizing antibodies, FRNA is a quantitative method that counts the individual cells that are initially infected with the virus and do not require several viral replication rounds. Each sample dilution tested can count more than 4,000 cells compared to about a few hundred for other assays. However, the method uses imaging instrumentation that may not be readily available in all labs.
They found a nucleocapsid protein antibody for SARS-CoV-1 that also had cross-reactivity with SARS-CoV-2, which the team used for the immunofluorescence staining.
After initial testing of samples, they found that there was no difference in the results between serum or plasma, so they used plasma for further studies. Their statistical analysis allowed them to determine acceptable values of the cutoff for greater accuracy in detection.
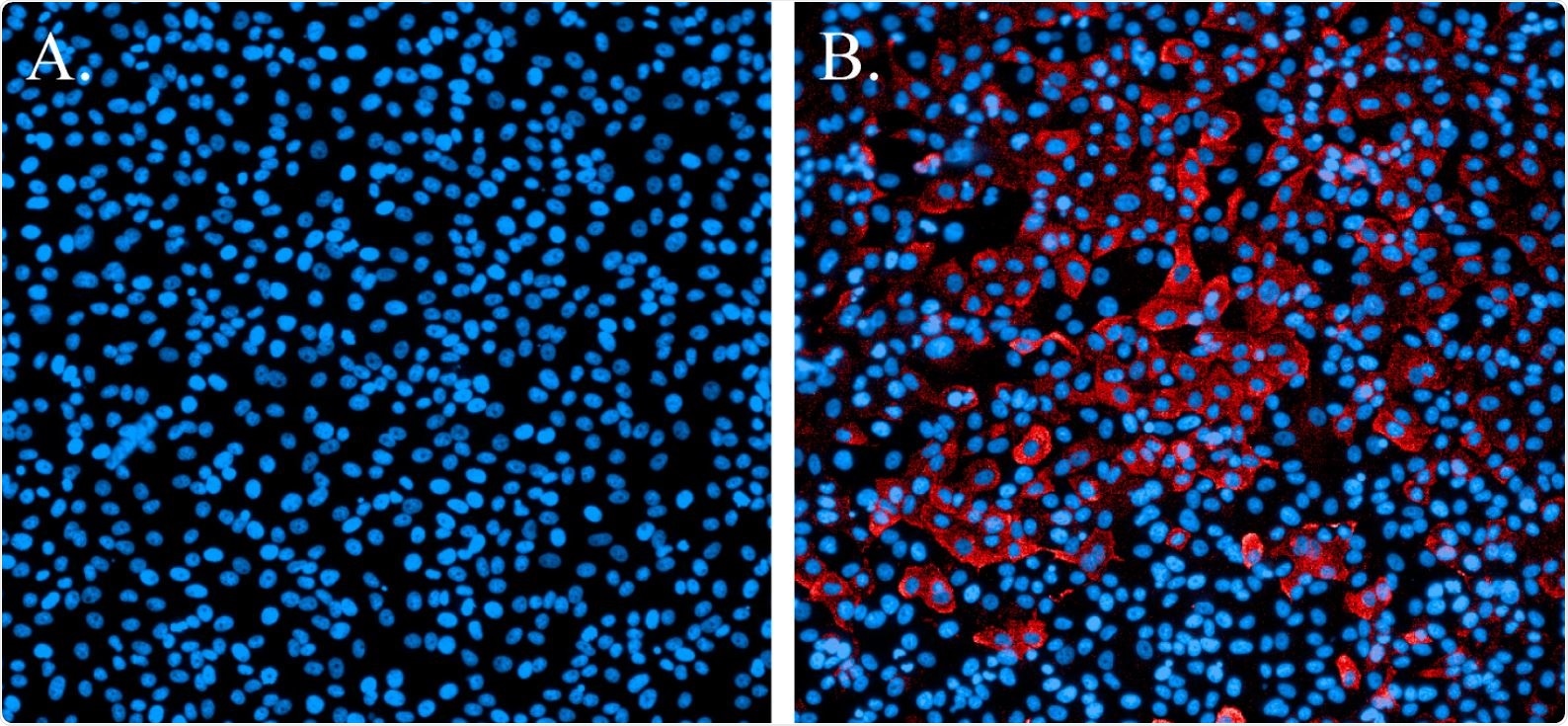
Immunofluorescence staining of SARS-CoV-2-infected cells. (A). Non-infected cells stained with Hoechst nuclear stain (blue). (B). Cells infected with SARS-CoV-2 and probed with a SARS-CoV N-protein-specific antibody and Alexa594 secondary antibody (red). Cells were counterstained with Hoechst nuclear stain (blue).
Quantitative results
When more than three values per plate were outside the cutoff, the entire experimental run was discarded. Based on an algorithm, they determined that only one run out of 15 was inaccurate. The rigorous statistical analysis thus allows confidence in the accuracy of the tests.
To determine how specific their assay was, the team compared their assay with total IgG obtained from two commercial ELISA assays. They found good agreement between the 50% neutralization titer (NT50) of the FRNA method and the ELISA positivity. There was some variability, however, in samples that were weakly positive by FRNA NT50. This was likely because of the high dilution of the samples. Less dilute samples may be able to detect borderline positive samples.
Generally, neutralization assays are not used as diagnostic tools, but the authors showed that their test correlated well with the diagnostic ELISA assay. Although the authors used the Operetta high-content imaging system, other similar equipment could be used after validation. Without the availability of a high-content imaging system, the method can be run as an immunofocus assay, but it will lose much of its power and throughput.
Assays involving live viruses are inevitable to have some variability, but using static analysis in this case can significantly reduce the errors introduced by inefficient infection, pipetting or staining errors.