As vaccine rollouts against the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus behind the coronavirus disease (COVID-19) pandemic, new variants of the virus threaten vaccine efficacy.
In the last quarter of 2020, variants, namely the B.1.1.7 and the B.1.351 variants, emerged in the United Kingdom and South Africa, respectively. In January 2021, a new variant, called the P.1. variant, emerged in travelers from Brazil. Recently, India reports another variant that has caused its skyrocketing cases, called the B.1.617 variant.
Now, researchers at the Infectious Diseases Unit, Rochester General Hospital in New York, report a SARS-CoV-2 B.1.1.345 variant carrying the E484K mutation in four patients with no apparent epidemiological associations in a hospital network in upstate New York.
The report, published in the journal Clinical Infectious Diseases, revealed that the variant detected has been spreading at an alarming rate since November. By February 2021, it accounted for 12.3 percent of all cases in the area.
Variants in the United States
The United States reports the highest number of COVID-19 cases, topping 33.38 million. In the past months, the country has detected new SARS-CoV-2 variants, with 8,337 cases with the B.1.1.7 variant identified in 51 jurisdictions, 266 cases with the B.1.351 variant, and 79 cases with the P.1 variant.
Scientists have also identified several regional variants, including the B.1.2 lineage carrying the Q677P mutation in New Mexico and Louisiana and the 20C-US lineage in the Midwest. However, these variants lack the E484K and the N501Y mutations responsible for making the virus spread faster and evade the immune system.
Recently, scientists have detected and described the B.1.536 variant in New York, which carries the E484K mutation. This mutation is not a new variant in itself but a mutation that occurs in different variants. The mutation is found in the spike protein and appears to impact the body’s immune response and vaccine efficacy.
Similar to previous studies about variants carrying the E484K mutation, antibody neutralization by the antibodies is impaired or stopped. Also, the neutralizing activities of convalescent plasma or post-vaccination blood were lower by 7.7-fold and 3.4-fold, respectively.
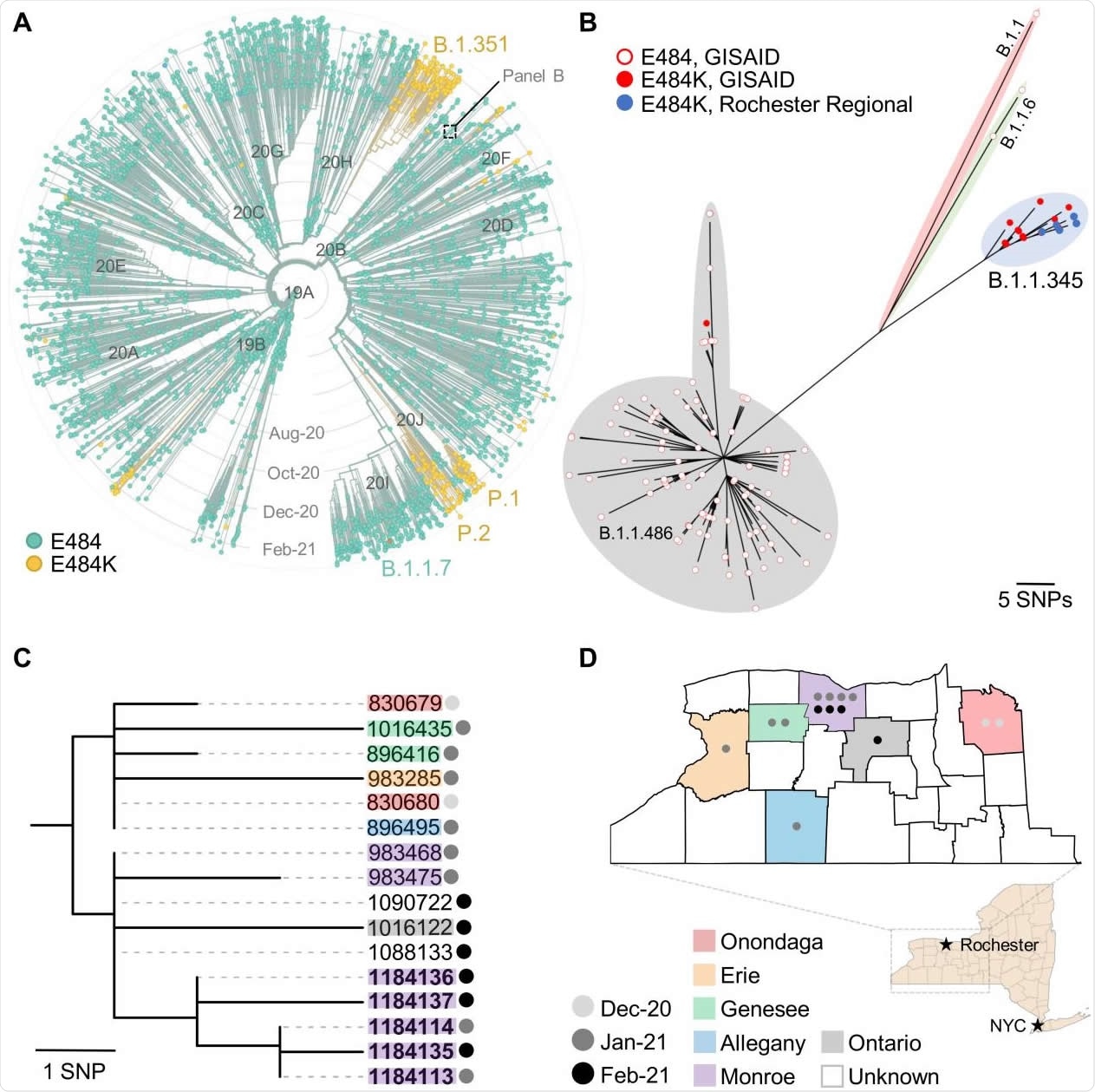
The emergence of the E484K mutation in B.1.1.345 SARS-CoV-2 isolates from Upstate New York. (A) Time-scaled, global phylogeny of 3,960 SARS-CoV-2 genomes sampled between Dec 2019 and Mar 2021 (modified from Nextstrain (https://nextstrain.org) dashboard). Major clades (Nextstrain nomenclature), variants (literature) and lineages (Pangolin) are indicated. Strains are colored based on the presence of a glutamic acid (E, teal) or lysine (K, 243 orange) at position 484 of the spike protein. (B) SNP-based, unrooted phylogram of B.1.1.345, B.1.1.486, B.1.1 and B.1.1.6 variants (colored shading) collected from New York State between October 2020 and March 2021 (n = 129). Empty circles indicate strains with Glutamic acid (E) at residue 484 of the spike protein while filled circles indicate those with Lysine (K) at that position. Genomes obtained from GISAID (www.gisaid.org) Genomes newly described in this report are shown in blue (note that only four out of five circles are visible as two samples from a single patient were identical). (C) SNP-based, mid-point rooted maximum likelihood phylogeny of sixteen monophyletic B.1.1.345 strains carrying the E484K mutation. Both comparator and newly sequenced isolates (bold font) are labeled using their GISAID Accession ID. Where known, the county (colored label) and collection date (colored circle) are indicated. For both panels B and C, the number of SNPs separating each strain can be inferred from the respective scale. (D) Map of the Finger Lakes region indicating the origin of fourteen B.1.1.345 strains carrying the E484K mutation (county of origin was not available for 1 isolate and the other is a duplicate sample from one patient). Inset map of the entire New York State and landmark cities are provided for geographical context.
The findings demonstrate the alarming rise of this new mutation, which has been found in 122 of 1,242 active lineages. When this mutation becomes rampant in many countries, it may threaten the current vaccination efforts to curb the pandemic.
A pilot study was started in June 2020 between the Multi-drug resistant organism Repository and Surveillance Network (MRSN) and the Rochester Regional Health System. The researchers extracted RNA from SARS-CoV-2-positive nasopharyngeal swabs and transferred it to the MRSN for sequencing.
In the most recent 20 samples collected from 19 hospitals between January 27 and February 7, 2021, the E484K mutation was detected in five samples collected from four patients. Three of these patients developed mild disease with no specific anti-SARS-CoV-2 treatments. One patient received two treatments, Remdesivir and Dexamethasone, but did not require intensive care or intubation.
All the patients lived in separate assisted living facilities with no known connections, and the fourth patient lived in a home.
The researchers also conducted a whole-genome high-resolution single nucleotide polymorphism (SNP) based comparison to better understand the correlation between the 16 B.1.1.345 samples. They found that out of the five samples, two from the same patient were genetically identical, and the remaining three samples were separated by 1-1-4 SNPs.
About 90 samples were collected from New York and adjacent countries, with the remaining being collected across the New York states. One sample collected on February 18 carried the E484K mutation, indicating that the mutation has emerged independently in B.1.1.486 in New York.
It is important to determine how the E484K mutation affects vaccine protection against the B.1.1.345 variant that lacks the other mutations seen in the B.1.351 and P1 strains.
“As the vaccine rollout gathers pace across the United States, every means necessary must be employed to rapidly identify the emergence of variants-of-concern and immediate action taken to limit their spread,” the researchers emphasized.