The humoral arm of innate immunity consists of numerous pattern recognition molecules, such as mannose-binding lectin (MBL) and pentraxin 3 (PTX3).
Humoral pattern recognition molecules persist as the ancestors of antibodies deployed in adaptive immune response, recognizing pathogens and eliminating them by a diverse range of mechanisms.
Generally, they bind with virus envelope glycoproteins, as has been observed in influenza, hepatitis C, and herpes simplex virus, inducing one of the several elimination routes available or directly preventing interaction with host cell membranes.
The most severe symptoms of COVID-19 are associated with an over-response of the cellular innate immune system, where a signaling cascade induces a cytokine storm, and our understanding of this aspect of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection has advanced considerably since the early stages of the pandemic.
There has, however, been less investigation into the role of the humoral arm of the innate immune system.
In a paper recently uploaded to the preprint server medRxiv* by Garlanda et al. (June 8th, 2021), humoral pattern recognition molecules are explored for affinity towards SARS-CoV-2, contributing to unraveling the role of these molecules in COVID-19 disease progression.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Identifying relevant pattern recognition molecules
A solid-phase binding assay was utilized to investigate any interaction between the ten selected humoral innate immunity molecules and SARS-CoV-2 proteins, specifically the: spike protein trimer, S1 and S2 subunits of the spike protein, nucleocapsid, and envelope proteins. Protein Coding gene PTX3 was seen to bind in a concentration-dependent manner to the nucleocapsid protein, and as this is a large glycoprotein with distinct C- and N-terminal domains, each domain was also tested against the nucleocapsid protein, finding that the N-terminal domain was the main contributor, though bound more weakly than the full-length protein.
Instead of the nucleocapsid, MBL was observed to bind with the spike protein of SARS-CoV-2, though not the separate subunits, possibly indicating that the closed form of the spike trimer is needed for interaction.
MBL is a member of the collectin family, bearing a Ca2+ type lectin domain, and so the group selected six additional molecules from this family for further analysis. None showed any affinity towards the spike protein compared with MBL, suggesting that the interaction is unique to this molecule and not due to the structural similarities shared by the collectin family.
The group modified a lentivirus vector to bear the SARS-CoV-2 spike protein and exposed to MBL, demonstrating specific interaction between the pseudovirus particles and the pattern recognition molecule.
As discussed, pattern recognition molecules largely interact with glycans, which heavily coat the surface of the SARS-CoV-2 spike protein. The protein bears 22 N-glycosylation sites, 8 of which contain oligomannose-type glycans that are probable interaction sites with the MBL carbohydrate recognition domain. The group exposed the spike protein to D-mannose or N-acetyl-glucosamine before MBL, finding that binding was now inhibited, and confirming the Ca2+ dependent pathway. The group searched the molecule for suitable binding sites, selecting for those with high oligomannosylation occupancy, and eventually identified two probable sites that span the S1 and S2 subunits, explaining the lack of interaction with them individually. The spike proteins of variants of concern B.1.1.7, B.1.1.28, and B.1.351 were also analyzed in silico for changes in possible binding sites with MBL, with the mutations not appearing to significantly alter the glycosylation sites. Docking simulations and in vitro testing using variant of concern spike proteins confirmed that MBL binds with an equal affinity towards wildtype and later lineages of SARS-CoV-2.
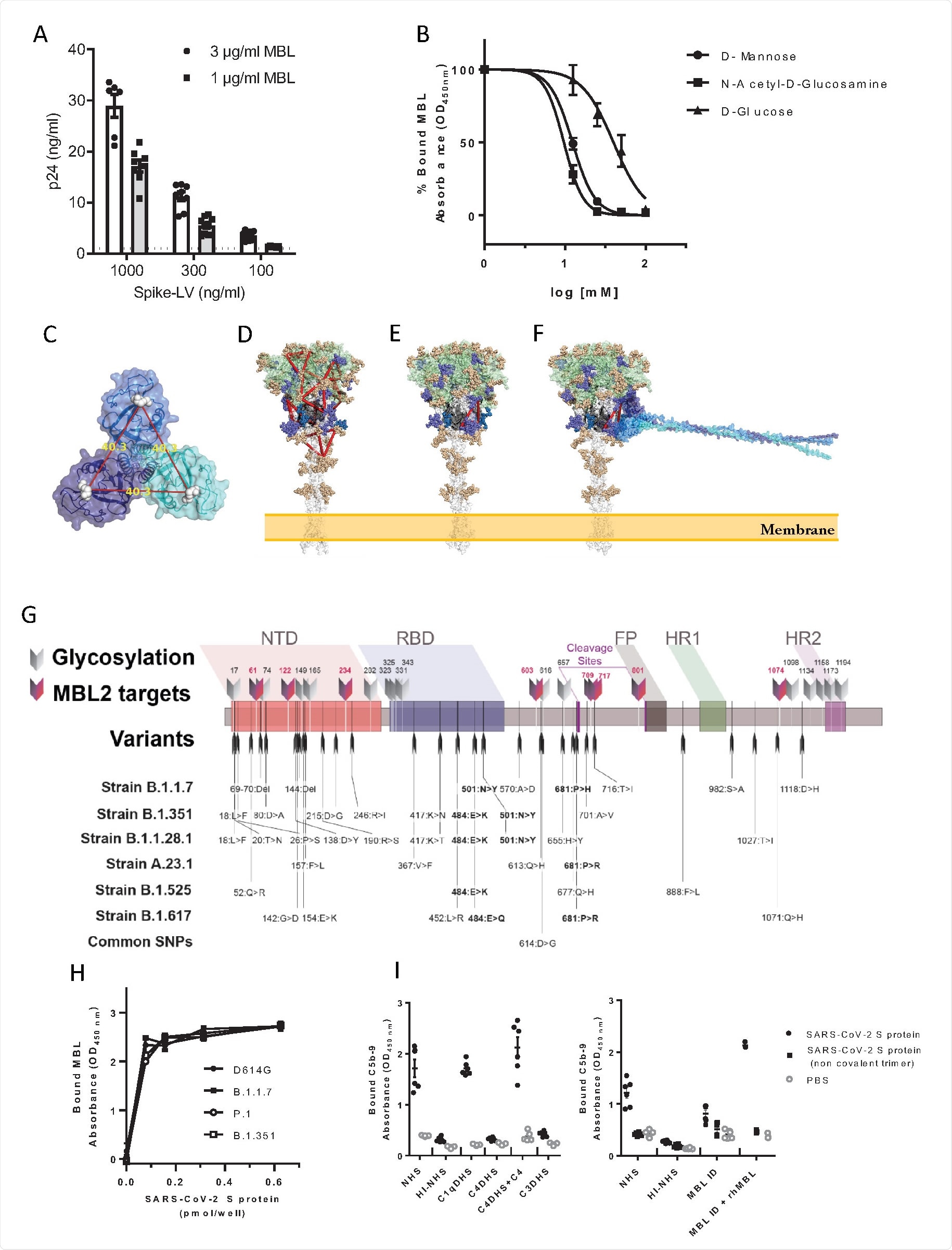
Interaction of MBL with SARS-CoV-2 S protein through its Carbohydrate Recognition Domain (CRD). (A) MBL-coated plates were incubated with different concentrations of viral particles of SARS-CoV-2 Spike protein (Spike-LV) or VSV g pseudotyped on a lentivirus vector. After lysis, bound pseudovirus was quantified by measuring the released p24 core protein by ELISA. Dotted line represents p24 levels measured using the highest concentration of VSV-g pseudotyped lentivirus. Data are represented as mean ± SEM of two independent experiments (n=6-10). (B) Biotinylated SARS-CoV-2 S protein was captured on Neutravidin coated plates. MBL (0.25 µg/mL) was incubated over S protein, alone or in the presence of different concentrations of D-mannose or N-acetyl-glucosamine or D-glucose. Binding of free MBL was detected by ELISA. Data are presented as percentage of bound MBL (mean ± SEM of two independent experiments, n=4). (C) Trimeric MBL2 model shows the distance of approximately 40 Å between mannose-binding sites. MBL2 subunits are represented in transparent surface and cartoon, colored in cyan, blue, and dark blue; mannose molecules are represented in white spheres; (D) 14 mannose-binding sites, represented as red triangles, imposed onto Spike protein. Spike is represented in transparent surface and cartoon, where S1 region (1-685) is colored in green, beginning of S2 region (686-815) is colored in black, and S2’ region is colored in white. (E) Putative binding site of MBL2. Pose with the highest site-specific probability to be glycosylated with oligomannose; (F) Spike-MBL2 complex. Glycosylation sites are colored according to the oligomannose glycosylation probability. Gold < 60%. Purple > 80% up until S2’ region. Blue > 80% in the S2’ region. (G) Schematic representation of glycosylation sites and nucleotide substitutions in the variant strains identified to date. The 8 glycosylation sites containing oligomannose-types glycans, which are potential targets of MBL, are evidenced. SNPs common to all variants are in bold. NTD: N terminal domain; RBD: receptor binding domain; FP: fusion peptide; HR1 and 2: heptad repeat 1 and 2; cleavage sites are reported. (H) Binding of MBL to SARS-CoV-2 S protein variants: recombinant SARS-CoV-2 S protein (D614G), B.1.1.7, P.1, and B.1.351 variants were captured onto a Nickel coated plate at different concentrations. MBL (1 µg/mL – 3.4 nM) was incubated over the captured viral proteins. Data are represented as mean ± SEM, n=3 replicates. One representative experiment out of two performed is shown for B.1.1.7. For the other variants, one experiment was performed. (I) MAC (C5b-9) deposition on SARS-CoV-2 Spike protein-coated plates. The assay was performed in the presence of 10% normal human serum (NHS), C1qDHS, C4DHS, C4DHS reconstituted with C4, C3DHS, MBL-ID serum, MBL-ID serum reconstituted with rhMBL. Mean ± SEM of two independent experiments, performed in triplicate (n=6).
Inhibition of SARS-CoV-2
To further test whether MBL inhibited cell entry by SARS-CoV-2, the spike protein-bearing lentiviral vectors were applied to Calu-3 cells and 293T cells heavily expressing the ACE2 receptor, with the pattern recognition molecule present at varying concentrations. At the highest tested concentration of 34 nM viral cell entry was inhibited by 90% in 293T cells, with an EC50 of around 1.7 nM. Calu-3 cells are lung-derived epithelial cells, and thus more relevant to the investigation of SARS-CoV-2 inhibitors, and against these cells, the calculated EC50 was just 0.27 nM at 72 hours post-infection.
MBL and PTX3 were also applied to a 3D human bronchial epithelial cell model, finding that the former was able to decrease viral production by an order of magnitude when applied at a concentration of 170 nM by 72 hours post-infection. The production of interleukin-8 and other key inflammatory cytokines was also inhibited by MBL, though presumably due to the lower rate of infection in tissues treated with the molecule.
Finally, the group performed data analysis of Italian COVID-19 patients where data regarding genetic variation in the MBL producing gene was available. One polymorphism, in particular, was highlighted as being associated with increased susceptibility to COVID-19 infection and more severe symptoms, which has also been implicated with a poorer prognosis in influenza and other respiratory infections.
The group suggests that recognition of SARS-CoV-2 by MBL could play both a positive and negative role in disease progression, as it serves as a form of antiviral in the early stages of infection, though it may then exacerbate the inflammatory response observed later in severe cases.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Recognition and inhibition of SARS-CoV-2 by humoral innate immunity pattern recognition molecules, Matteo Stravalaci, Isabel Pagani, Elvezia Maria Paraboschi, Mattia Pedotti, Andrea Doni, Francesco Scavello, Sarah N. Mapelli, Marina Sironi, Luca Varani, Milos Matkovic, Andrea Cavalli, Daniela Cesana, Pierangela Gallina, Nicoletta Pedemonte, Valeria Capurro, Nicola Clementi, Nicasio Mancini, Pietro Invernizzi, Rino Rappuoli, Stefano Duga, Barbara Bottazzi, Mariagrazia Uguccioni, Rosanna Asselta, Elisa Vicenzi, Alberto Mantovani, Cecilia Garlanda, medRxiv, 2021.06.07.21258350; doi: https://doi.org/10.1101/2021.06.07.21258350, https://www.medrxiv.org/content/10.1101/2021.06.07.21258350v1
- Peer reviewed and published scientific report.
Stravalaci, Matteo, Isabel Pagani, Elvezia Maria Paraboschi, Mattia Pedotti, Andrea Doni, Francesco Scavello, Sarah N. Mapelli, et al. 2022. “Recognition and Inhibition of SARS-CoV-2 by Humoral Innate Immunity Pattern Recognition Molecules.” Nature Immunology 23 (2): 275–86. https://doi.org/10.1038/s41590-021-01114-w. https://www.nature.com/articles/s41590-021-01114-w.