Researchers have found that the variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that were initially identified in South Africa (B.1.351) and Brazil (P.1) appear to be spreading more quickly in some areas of France than the previously dominant UK variant B.1.1.7.
The SARS-CoV-2 virus is the agent responsible for the ongoing coronavirus disease 2019 (COVID-19) pandemic that has now claimed the lives of more than 3.8 million people globally.
The team found that since the period between January and March 2021, when the transmission advantage of B.1.1.7 was larger than that of B.1351, that trend appears to have shifted in at least two French regions during April. The B.1.351 and P.1 lineages showed a significant transmission advantage over B.1.1.7 in the regions between April 12th and May 7th.
The researchers say that since one of these regions – Île-de-France – has been one of the most impacted by the epidemic to date, it is possible that a shift in variants with a transmission advantage is occurring there due to the high proportion of individuals who have acquired immunity to SARS-CoV-2 through previous infection.
The team from Cerba Laboratory in Saint Ouen L’Aumône, Montpellier University Hospital and the University of Montpellier says that, given the ongoing relaxation of control measures in June, the findings call for particular care to be taken regarding vaccination rollout and the maintenance of non-pharmaceutical interventions until vaccine coverage reaches levels that are compatible with spontaneous regression of the epidemic.
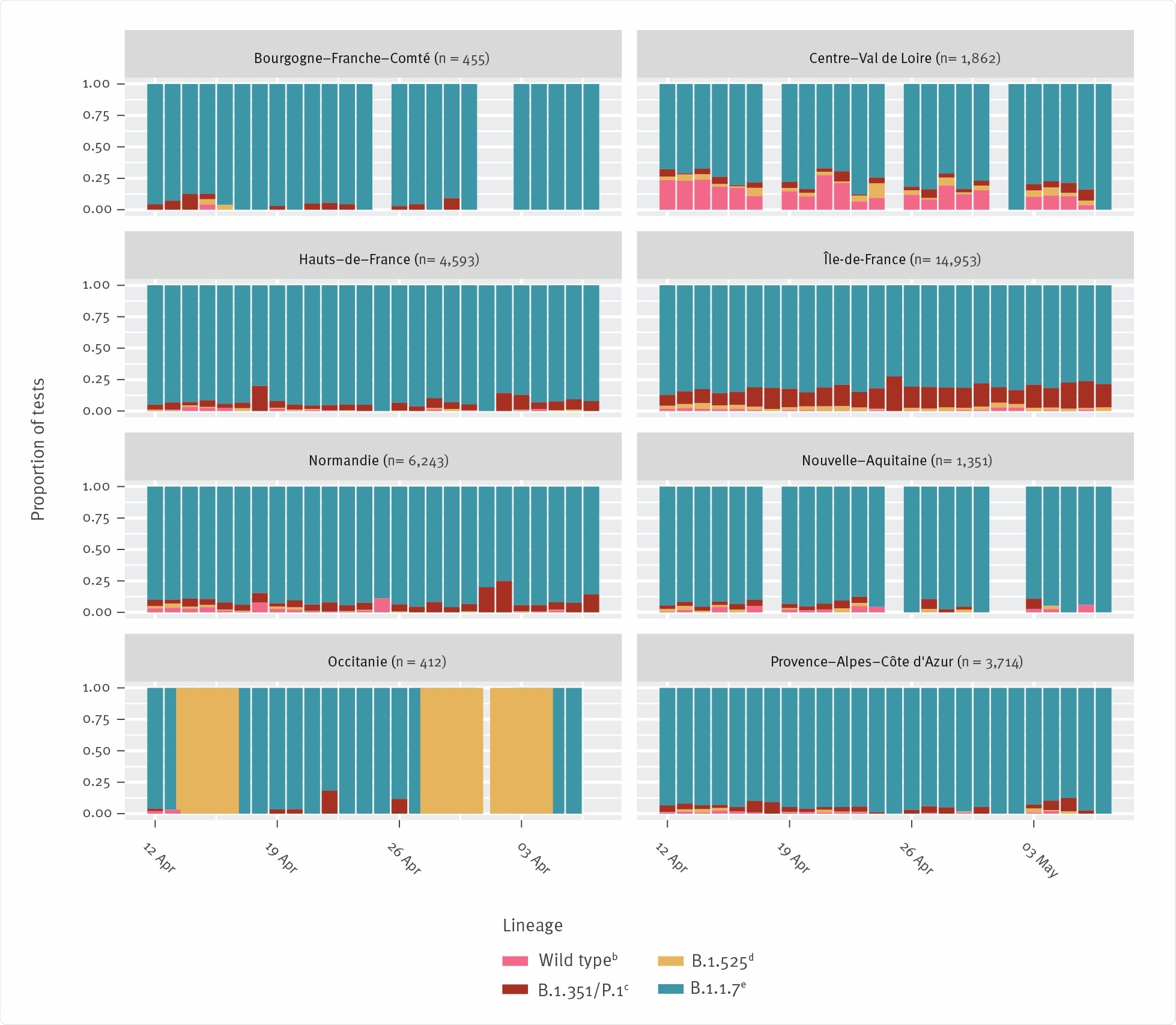
SARS-CoV-2: severe acute respiratory syndrome coronavirus 2; VOC: variant of concern; VOI: variant of interest. a Only regions with more than 400 respective tests are shown. b Characterisation as wild type SARS-CoV-2 is based on the absence of both N501Y and E484K mutations. c Characterisation as B.1.351 and/or P.1 lineage (VOC β and/or γ) is based on the presence of both N501Y and E484K mutations. d Characterisation as B.1.525 lineage (VOI η) is based on the simultaneous absence of N501Y and presence of E484K mutation. e Characterisation as B.1.1.7 lineage (VOC α) is based on the simultaneous presence of N501Y and absence of E484K mutation. The number of tests performed is indicated in each panel. For each day, the different colours indicate the proportion of tests belonging to each of the four screening categories (these sum to 1.0 every day). Regions with few tests can exhibit strong variations in frequencies for some days (e.g. Occitanie for days with only B.1.525 detected).
More about the variants of concern so far identified
The SARS-CoV-2 variants of concerns (VOCs) that have emerged during the COVID-19 pandemic are phenotypically distinct lineages associated with major epidemiological or clinical shifts.
To date, four viral lineages have been classified as VOCs by the World Health Organization. The first emerged in the UK (B.1.1.7) and is currently causing the majority of infections in Europe and North America. The second emerged in South Africa (B.1.351), where it is currently the most common strain. The third variant – P.1 – dominates in Brazil and South America, and the fourth – B.1.617.2 – caused a major epidemic wave in India.
Since January 2021, national guidelines have required that all clinical samples testing positive for SARS-CoV-2 undergo additional reverse-transcription polymerase chain reaction (RT-PCR) testing to detect mutations indicative of certain variants.
Since April 2021, this variant-specific testing has targeted the N501Y mutation that is shared by the B.1.1.7, B.1.351 and P.1 lineages and the E484K mutation shared by B.1.351, P.1, and the B.1.525 variant of interest (VOI) that emerged in Denmark.
What did the current study involve?
To investigate the transmission of SARS-CoV-2 variants in France, Samuel Alizon and colleagues analyzed the results of 36,590 variant-specific RT-PCR tests that were performed on samples between April 12th and May 7th, 2021, in 13 regions of the country.
As reported in the journal Eurosurveillance, the dominant lineage in most regions was B.1.1.7, which made up 79.1% of viruses detected.
Other prevalent lineages included B.1.351 (7.9%,), B.1.525 (4.4%) and B.1.214 (2.3%). The latter, which was first identified in Switzerland, is not yet classified as a VOC, but is currently undergoing monitoring. Other lineages made up less than 2% of the samples, including P.1 (0.6%).
The B.1.351 and P.1 lineages were most frequently detected in the Île-de-France and Hauts-de-France regions.
For all regions, the risk of being infected with a wildtype or B.1.525 variant were either the same or lower than the risk of being infected with B.1.1.7.
However, the risk of being infected by B.1.351 or P.1 rather than B.1.1.7 significantly increased over time in Île-de France and Hauts-de-France.
For B.1.351 and P.1, the team identified a transmission advantage over B.1.1.7 of 15.8% in Île-de-France and 17.3% in Hauts-de-France.
How does this compare to findings earlier in the year?
The team had previously analyzed the results of variant-specific tests on samples obtained between January and March 2021. That study revealed that B.1.1.7’s transmission advantage over wildtype lineages was more significant than that of B.1.351.
“During April 2021, in at least two French regions, this trend appears to have shifted, with B.1.351 and possibly P.1 spreading more rapidly than B.1.1.7,” says Alizon and colleagues.
The researchers point out that the B.1.351 lineage has already been shown to escape vaccine- or infection-induced immunity.
“With Île-de-France being one of the French regions the most impacted to date by the epidemic, it is possible that a shift in variants with a transmission advantage is occurring there because of the high proportion of individuals with immunity acquired through prior-SARS-CoV-2-infections,” they suggest.
More detailed analyses are needed
The team also points out that the coverage resulting from the COVID-19 vaccination rollout is homogeneous among French regions.
Alizon and colleagues say the findings call for more detailed analyses of the link between the transmission advantage of B.1.351 and the proportion of the population that is immune to SARS-CoV-2 (following infection or vaccination) in different French regions.
“Given the progressive lifting of the control measures in June 2021 in France, these results call for particular care regarding vaccination rollout and the maintenance of non-pharmaceutical prevention until vaccine coverage reaches levels compatible with spontaneous regression of the epidemic,” they conclude.