The alpha variant (B.1.1.7) of severe acute respiratory syndrome coronavirus -2 (SARS-CoV-2) emerged as the dominant strain in the UK in late 2020 and early 2021. Infectivity is enhanced, immunity is compromised, and clinical outcomes are more severe in variants with mutations in the spike protein.
A total of 253 positive samples were selected for this study, all of which were collected from December to February 2021. Besides the qualitative RT-PCR results, the threshold Ct value and patient metadata (age, sex, etc.) were considered in the study. Nasopharyngeal swab (NPS) samples were processed as per coronavirus disease - 19 (COVID-19) diagnostic protocols and standard operating procedures.
Subsequently, diagnostic RNA extraction and RT-PCR were carried out. Random positive samples were processed for full viral genome sequencing. The research was undertaken by the University of Birmingham’s facility of COVID-19 UK Genomics UK Consortium (COG-UK).
The next step was to investigate the presence of N501Y and ΔH69/V70 mutations, and this was performed by probe-based melting curve one-step RT-PCR assays, using VirSNiP mutation assays by TibMol (#53-0780 and #53-0781, TIB Molbiol, Berlin, Germany).
Data on biomarkers were obtained from hospitals and were anonymized. The number of hospital admissions and outcomes over time and average values of specific biomarkers over the same period and two months prior were analyzed, from 1,112 blood specimens.
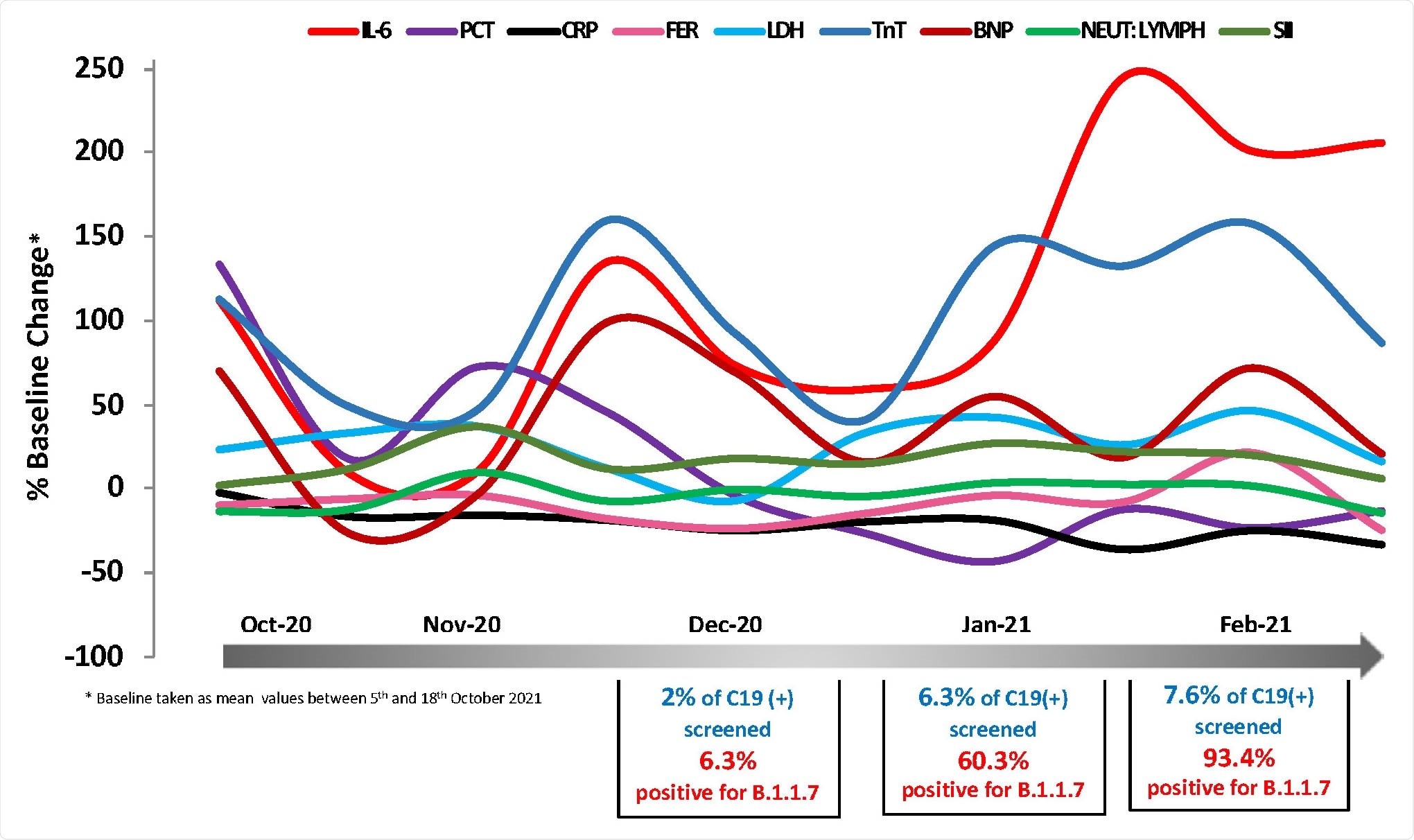
Temporal changes in blood COVID-19 biomarkers during the period 5/10/2020- 7/3/2021. Mean values expressed as % change from baseline (mean value of 5t -18th October 2020) are plotted as a function of time using measurements at regular time intervals (twice a month). Biomarkers of interest were measured at the routine Biochemistry laboratory of UHCW NHS Trust using CE-marked Elecsys kits from Roche Diagnostics. In total 1,112 data points were analyzed.

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
An independent t-test was used to compare biomarker values between groups. Where available, the B.1.1.7 status and associated biochemistry parameters were compared across groups to gauge any differences across samples. Melting temperature shifts of the fluorescence peak from 55.9 - 66.5 degrees Celsius (+/- 2.5) indicate the presence of the N501Y mutation. The melting temperature shift was 56.5 – 63.4 degrees Celsius for the ΔH69/V70 mutation.
Both assays exhibited robust reproducibility and repeatability characteristics and a wide range of primary RNA samples with Ct values ranging from 15 to 28 were analyzed. Out of the 253 RNA samples, successful implementation of the PCR protocol was achieved in 235 samples. The N501Y mutation assay had a failure rate of around 7%, whereas the failure rate of the ΔH69/V70 assay was 2%.
A considerable advantage of the method used is the short reaction time of each RT-PCR amplification (less than 1 hour), enabling the generation of results on the same day. It was observed that all RNA samples that tested positive for the N501Y were also positive for the ΔH69/V70 mutation.
After identification of the alpha strain, researchers analyzed its impact on various biomarkers. It was noted that three biomarkers, namely, IL-6, BNP, and cTnT showed a transient increase during the first half of December 2020 and, subsequently, a continued rise during the second half of January and February 2021. Levels of IL-6 were disproportionately high. Similar trends were demonstrated by markers of cardiac function cTroponin T (cTnT) and NT-pro-BNP, albeit not significant. Other inflammatory markers such as CRP, ferritin, and LDH did not demonstrate an identical pattern.
Researchers of the current study used their data to analyze the biomarker levels in hospitalized patients with or without the B.1.1.7/alpha variant of concern. They focused on a small sample of patients (44 patients tested positive for the alpha variant while 39 tested negative) for whom biomarkers and clinical outcome data were available. The survivor-deceased ratio was 3:1 in the alpha variant positive group, while it was 1:1 in the negative group. Patients were grouped according to their outcome (survived or deceased) and the alpha variant being positive or negative.
For both the groups, deceased patients showed higher modified z-scores for several biomarkers, whereas survivors had low or negative z-scores. As mentioned earlier and as expected, IL-6 demonstrated the most marked difference in alpha variant positive deceased patients vs. survivors, followed by LDH. There was a significant difference in the modified z-scores of most inflammatory biomarkers in both the alpha positive and negative groups. These include CRP, N/L ratio, pro-NTBNP and cTnT.
These results hint towards a higher and more potent inflammatory response in the deceased patients and also the activation of multiple biomarkers and pathogenic pathways. LDH and SII levels were higher than the overall group median for patients who tested positive for the alpha variant, irrespective of the clinical outcome.
In summary, mutation detection assays can be suitable to detect SARS-CoV-2 variants. Rapid deployment of such tools can aid in preventing transmission of variants of concern and help contain the ongoing COVID-19 pandemic.

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.