Researchers have conducted a study showing that the B.1.1.7 lineage of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that emerged in the UK is now rapidly being displaced as the dominant strain in the United States by the variants of concern B.1.617.2 (Delta) and P.1 (Gamma) that emerged in India and Brazil, respectively.
The SARS-CoV-2 virus is the agent responsible for the ongoing COVID-19 pandemic that has now claimed the lives of more than 3.83 million people globally.
Since B.1.617.2 has displaced B.1.1.7 as the dominant variant in England and other countries, Alexandre Bolze and colleagues from Helix in San Mateo, California, set out to determine whether it is also displacing B.1.1.7 in the United States.
The team’s analysis of polymerase chain reaction (PCR) and viral sequencing results collected from across the United States showed that B.1.1.7 is no longer responsible for the majority of new cases in the country.
The percentage of SARS-CoV-2-positive cases that were of the B.1.1.7 lineage dropped from 70% in April 2021 to 42% over a period of just 6 weeks.
“Our analysis showed rapid growth of variants B.1.617.2 and P.1 as the primary drivers for this displacement,” says the team.
The study also showed that B.1.617.2 had a higher growth rate than P.1 and was growing faster in counties with a lower vaccination rate.
A pre-print version of the research paper is available on the medRxiv* server, while the article undergoes peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The concerns surrounding B.1.617.2
The B.1.617.2 lineage of SARS-CoV-2, which was recently classified as a variant of concern, has displaced B.1.1.7 as the dominant variant in England and other countries.
Moreover, while a study by Public Health England showed that full immunization (two doses) with the AstraZeneca or Pfizer-BioNTech vaccine remains more than 90% effective at protecting against hospitalization following infection with B.1.617.2, the efficacy following one dose is lower, compared with B.1.1.7 infection, says Bolze and colleagues.
The B.1.617.2 variant was first identified in the United States on March 16th, 2021.
However, the United States differs from England in terms of vaccine strategies and the existing viral background, say the researchers.
In England, B.1.1.7 represented more than 90% of SARS-CoV-2 sequences when B.1.617.2 was first identified and there were very few sequences of P.1.
“In the United States, B.1.1.7 plateaued just above 70%, and there was a greater diversity of variants when B.1.617.2 started to emerge, including an increasing amount of P.1,” writes the team.
What did the researchers do?
To examine the impact that the B.1.617.2 and P.1 variants of concern have had on the prevalence of B.1.1.7 in the United States, the researchers analyzed PCR and sequencing results of samples collected by the Helix laboratory across the country since April 2021.
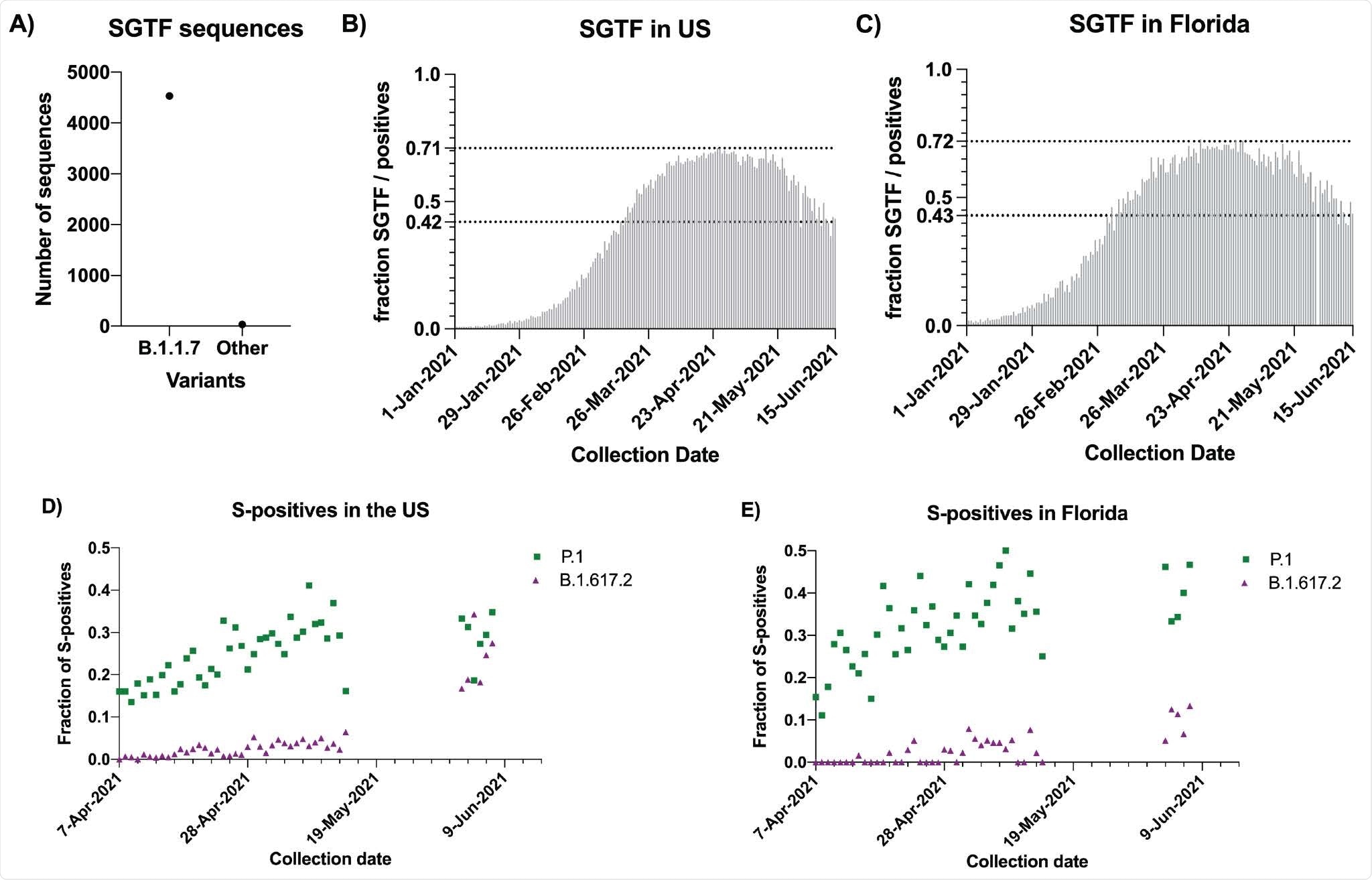
B.1.1.7 is being replaced by B.1.617.2 and P.1 in the United States. A) Counts of S-Gene Target Failure (SGTF) sequenced in May and June 2021 that were B.1.1.7 or Other variants. B) Fraction of SGTF to total positives per day in the US. The two dotted lines correspond to the maximum level observed in the US (71%) and the current level (42%). C) Fraction of SGTF to total positives per day in Florida. The two dotted lines correspond to the maximum level observed in Florida (72%) and the current level (43%). D) Fraction of S-positives sequenced that were either B.1.617.2 (purple triangles) or P.1 (green squares) by day in the US. No sequences were available from May 14 to June 2 2021. E) Fraction of S-positives sequenced that were either B.1.617.2 (purple triangles) or P.1 (green squares) by day in the US.
The Helix COVID-19 Test is based on the Thermo Fisher TaqPath COVID-19 Combo Kit, which targets three SARS-CoV-2 viral regions – the nucleocapsid (N) gene, the spike (S) gene, and open reading frame 1ab (ORFab1).
One defining mutation of the B.1.1.7 variant of concern is the deletion of amino acids 69 and 70 in the S protein.
“This deletion interferes with the PCR test target on the S gene in many COVID-19 tests, including the Helix COVID-19 Test, and causes S-gene target failure (SGTF),” explains Bolze and colleagues.
The team says SGTF is, therefore a reliable test for assessing the epidemiological dynamics of B.1.1.7.
To calculate the frequency of other viral variants, the team used sequencing data for non-SGTF samples.
The researchers also obtained data on the vaccination rate by county from the Centers for Disease Control and Prevention (CDC). They used the percentage of individuals completely vaccinated as of May 1st, 2021 for the analysis.
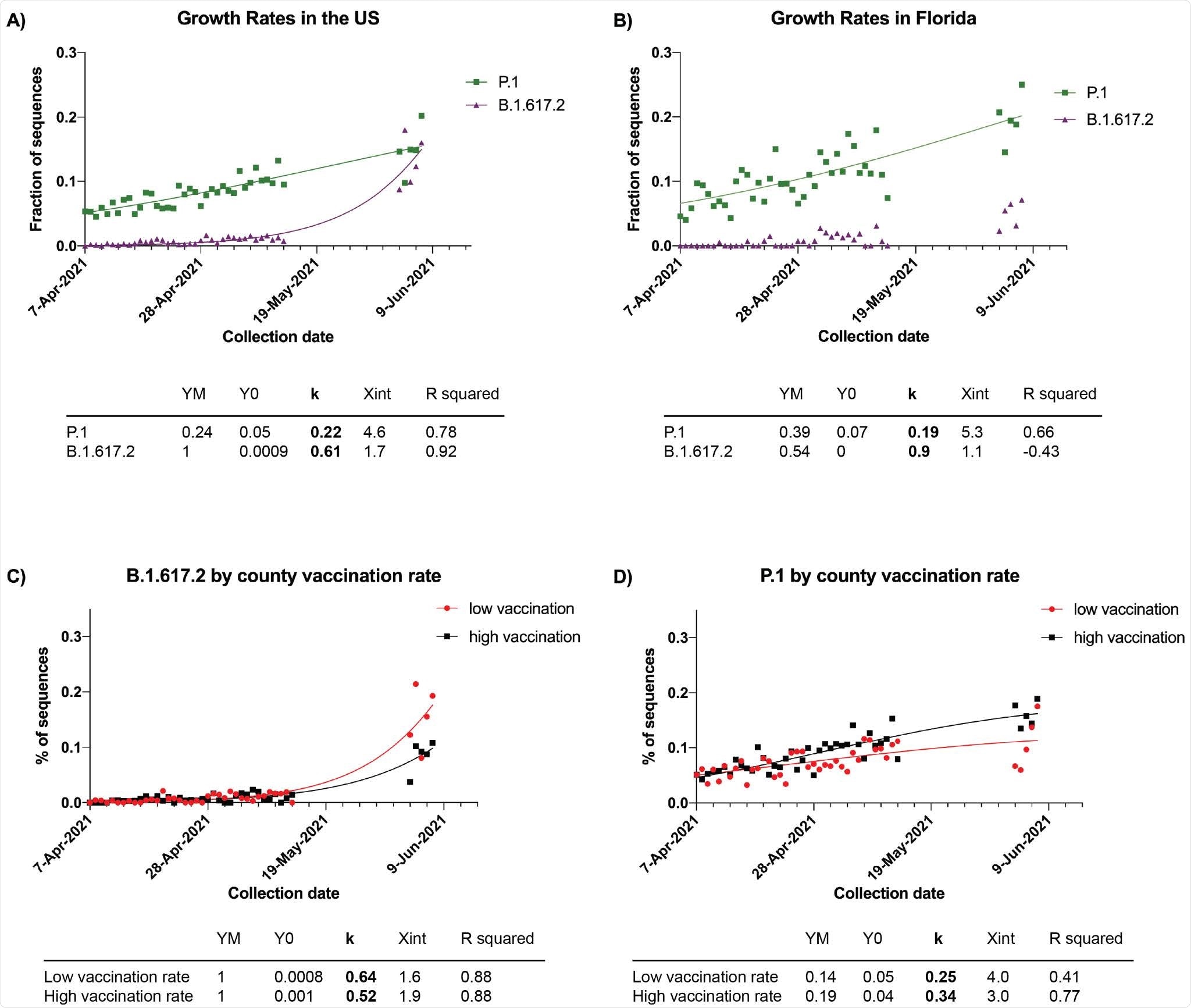
Growth rates of B.1.617.2 and P.1 in the United States. A) and B) Fractions of total sequences (SGTF or not) by day in the US (panel A) and in Florida (panel B) that were B.1.617.2 (purple triangle) or P.1 (green squares). A logistic growth curve was then fitted and is represented by the continuous purple line for B.1.617.2 and green line for P.1 The table below the graphs shows the key values of the curve Y=YM*Y0/((YM-Y0)*exp(-k*x) +Y0). YM is the maximum population; Y0 is the starting population; k is the rate constant. R squared is a measure of the goodness of fit. C) and D) Growth curves of B.1.617.2 (panel C) and P.1 (panel D) by county vaccination rate. Red represents counties with a low vaccination rate (below 28.5% completely vaccinated on May 1). Black represents counties with a high vaccination rate (above 28.5% completely vaccinated on May 1). Each symbol indicates the fraction of B.1.617.2 or P.1 to the number of samples sequenced per day. Characteristics of the curves are below each panel.
What did the study find?
Both SGTF and sequencing data indicated that after becoming the dominant SARS-CoV-2 lineage in the United States, the prevalence of B.1.1.7 plateaued at around 70% in April 2021.
The May and June test results pointed to a clear and rapid decrease in the prevalence of B.1.1.7, from 69.6% on April 26th-30th to 42% on June 11th-15th.
Sequencing of the non-SGTF (S-positive) samples collected between the 2nd and 8th June (when B.1.1.7 prevalence was rapidly decreasing) showed that P.1 represented 28% and B.1.617.2 represented 24.6% of the S-positive samples.
Analysis of the variants’ growth rates showed that both B.1.617.2 and P.1 were growing faster than B.1.1.7. However, B.1.617.2 is growing faster than P.1, at 0.61 versus 0.22, which the team says is consistent with reports from other countries.
Growth rates differed by county vaccination rate
Next, the researchers compared the Helix sequencing data by county with the county vaccination rates reported by the CDC.
This revealed that the prevalence of B.1.617.2, which is more transmissible but less resistant to vaccination, is higher in counties with lower vaccination rates. By contrast, P.1, which is less transmissible but more resistant to vaccination, is more prevalent in counties with higher vaccination rates.
The team expects that B.1.617.2 will soon become dominant
The researchers say they expect that B.1.617.2 will soon become the dominant variant in the United States.
“However, questions remain over whether it will entirely take over as it is doing in England, or whether it will plateau at a lower level like B.1.1.7 did in the US,” they add.
The team says B.1.617.2 may not reach as high levels in the United States due to the more diverse sets of policies between states regarding vaccinations and other public health measures.
“With this in mind, we showed that B.1.617.2 is growing more rapidly in counties with lower vaccination rates,” concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Bolze A, et al. Rapid displacement of SARS-CoV-2 variant B.1.1.7 by B.1.617.2 and P.1 in the United States. medRxiv, 2021. doi: https://doi.org/10.1101/2021.06.20.21259195, https://www.medrxiv.org/content/10.1101/2021.06.20.21259195v1
- Peer reviewed and published scientific report.
Bolze, Alexandre, Shishi Luo, Simon White, Elizabeth T. Cirulli, Dana Wyman, Andrew Dei Rossi, Henrique Machado, et al. 2022. “SARS-CoV-2 Variant Delta Rapidly Displaced Variant Alpha in the United States and Led to Higher Viral Loads.” Cell Reports. Medicine 3 (3): 100564. https://doi.org/10.1016/j.xcrm.2022.100564. https://www.cell.com/cell-reports-medicine/fulltext/S2666-3791(22)00071-4?