Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) testing along with contact tracing, isolation, and physical barriers like masks and distancing are the standard strategies used to reduce community transmission. However, insufficient testing protocols have led to failure in containing the community spread of SARS-CoV-2 in many countries. In addition, long delays in access to testing and obtaining test results have made it difficult to contain the spread of the virus. This gap in testing reduces the effectiveness of contact tracing and isolation strategies.
Despite the development of safe vaccines against the virus, the threat of transmission remains high due to the logistical difficulties of global vaccine distribution, limited supply of vaccines, and vaccine hesitancy. In addition to this, the emergence of more infectious SARS-CoV-2 variants that escape natural or vaccine-induced immunity shows that testing will remain a crucial tool in the fight against the pandemic to decrease viral transmission. Also, the absence of global vaccination coverage increases the potential for future outbreaks.
Frequent testing with ‘rapid’ tests that provide quick results is an effective strategy to monitor viral spread in the population and identify infectious individuals, thus allowing faster and safer reopening of the economy. An effective frequent testing strategy should be easy to use and self-administer, rapid, inexpensive, sensitive enough to detect most infectious people, and highly specific. Initial testing strategies for SARS-CoV-2 included deep nasopharyngeal swabs and RT-qPCR, and these tests were conducted by trained healthcare personnel.
To increase testing capacity and decrease time to test results, home-based diagnosis using ‘rapid’ tests were developed to detect viral antigens in shallow nasal swabs. While these tests offered quick results, they were not very sensitive and can only detect ~20,000 viral RNA copies per microliter.
A cheap, rapid, sensitive, and specific molecular test for the detection of SARS-CoV-2 in saliva
Researchers from the US recently described a rapid molecular test named WHotLAMP for the detection of SARS-CoV-2 in saliva. WHotLAMP is easy to use, very sensitive at 3.6 viral RNA copies per microliter of saliva, specific, and inexpensive. This study is published on the medRxiv*preprint serve.
WHotLAMP does not use toxic chemicals or special equipment and can be performed in point-of-care settings. WHotLAMP can also be adapted for home use. It can also be adapted for diagnostic assays for detecting other pathogens and is thus useful for potential future outbreaks.
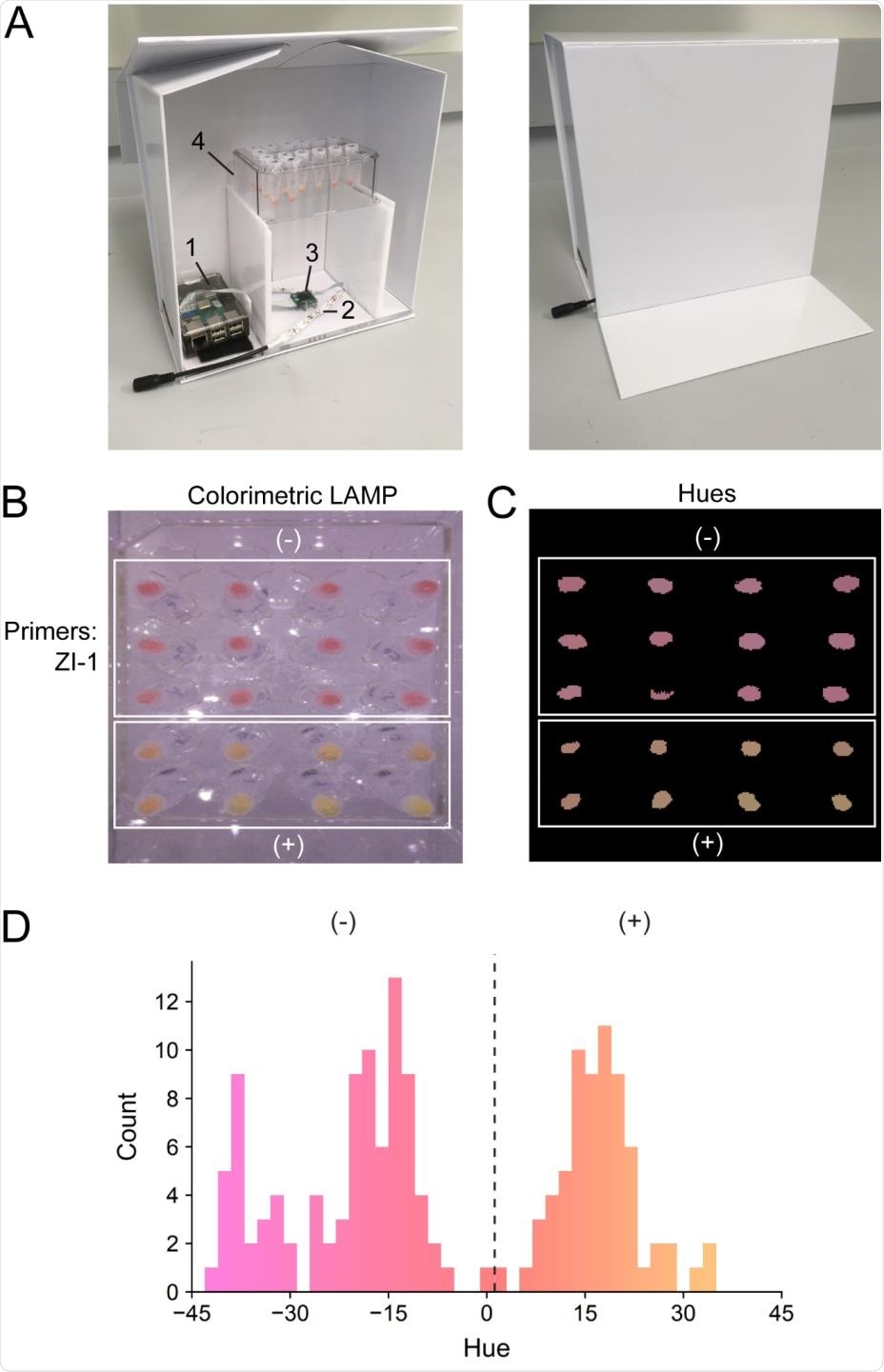
Colorimetric quantification of LAMP reactions. (A) Illuminated lightbox with automated image acquisition using Raspberry Pi. 1) Raspberry Pi unit; 2) white LED strip; 3) camera unit; 4) test tube rack. (B) LAMP reactions using WHotLAMP with ZI-1 primers on saliva samples from different negative nasal231 swab qPCR SARS-CoV-2 individuals (top white box) and SARS-CoV-2 positive (nasal swab) samples (bottom white box). (C) Processed image showing conversion of colorimetric LAMP results to hues. (D) Hue distribution of WHotLAMP saliva results from negative (-) and positive (+) nasal-swab SARS-CoV-2 qPCR donor samples.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
WHotLAMP is a single-tube test with primers highly specific to SARS-CoV-2
WHotLAMP testing can be performed in a single 1.7 mL microfuge tube. The viral RNA is preserved with the help of a non-hazardous RNA stabilizer and extracted through a brief Proteinase K digestion.
Heating the sample to 95 – 155oC inactivates Proteinase K as well as SARS-CoV-2 virions, thus increasing the biosafety of the saliva sample.
The researchers designed LAMP primer sets throughout the SARS-CoV-2 genome and targeted the primer set (ZI-1, targeting ORF 1a) with the lowest predicted propensity for primer-dimer formation. They also tested a panel of 22 inactivated respiratory pathogens such as SARS-CoV-1, MERS, H1N1 influenza, and common respiratory coronaviruses to evaluate the specificity of the ZI-1 primers. They detected SARS-CoV-2 RNA in samples with encapsulated SARS-CoV-2 RNA particles, but not in samples with only the other respiratory pathogens, illustrating the primers’ specificity to SARS-CoV-2.
WHotLAMP is a broadly useful diagnostic assay that can also be adapted to detect pathogens other than SARS-CoV-2
In this study, the authors describe WHotLAMP, a simple, affordable molecular test that does not need special lab equipment to detect SARS-CoV-2 in saliva. They show that WHotLAMP can detect low levels of SARS-CoV-2 virus present in saliva in about 30 minutes. Its low false-positive rate permits deployment in low prevalence conditions, where high test specificity is critical to obtain high positive predictive values. This assay design is applicable to point-of-care settings, and the single-tube format of the test requires no centrifugation, thus making it easy to scale to 96-well formats.
“While here we focused on a test for SARS-CoV-2, this technology could be used to detect other pathogens that are present in saliva by substituting primers, making WHotLAMP a broadly useful diagnostic assay.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
WHotLAMP: A simple, inexpensive, and sensitive molecular test for the detection of SARS-CoV-2 in saliva, David Ng, Ana Pinharanda, Merly C Vogt, Ashok Litwin-Kumar, Kyle N Stearns, Urvashi V Thopte, Enrico Cannavo, Armen Enikolopov, Felix Fiederling, Stylianos Kosmidis, Barbara Noro, Ines Rodrigues-Vaz, Hani J Shayya, Peter Andolfatto, Darcy S Peterka, Tanya Tabachnik, Jeanine D'Armiento, Monica P Goldklang, Andres Bendesky, medRxiv, 2021.06.17.21259050; doi: https://doi.org/10.1101/2021.06.17.21259050, https://www.medrxiv.org/content/10.1101/2021.06.17.21259050v2
- Peer reviewed and published scientific report.
Ng, David, Ana Pinharanda, Merly C. Vogt, Ashok Litwin-Kumar, Kyle Stearns, Urvashi Thopte, Enrico Cannavo, et al. 2021. “WHotLAMP: A Simple, Inexpensive, and Sensitive Molecular Test for the Detection of SARS-CoV-2 in Saliva.” Edited by Etsuro Ito. PLOS ONE 16 (9): e0257464. https://doi.org/10.1371/journal.pone.0257464. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0257464.