Researchers in the UK and Spain have used a novel drug screening approach to identify compounds that could serve as effective antivirals against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – the agent that causes coronavirus disease 2019 (COVID-19).
The team used a quantum-inspired device in combination with a more traditional fingerprinting method to search for drugs that are similar to remdesivir, the only antiviral against SARS-CoV-2 that is currently approved for human use.
“The COVID-19 pandemic has accelerated the need to identify new therapeutics at pace, including through drug repurposing,” says the team from Fujitsu Technology Solutions in Madrid and King’s College Hospital in London.
While both models predicted the antiviral drug GS-6620 as the top compound, the quantum-based model predicted the antiviral BMS-986094 as second best. Both of these compounds were initially developed to treat the hepatitis C virus.
The more traditional Tanimoto model also predicted different forms of vitamin B12 as potential antiviral candidates.
In vitro analyses revealed that BMS-986094 and the different types of B12 were effective at inhibiting the replication of SARS-CoV-2 variants.
“While BMS-986094 can cause secondary effects in humans as established by phase II trials, these findings suggest that vitamin B12 deserves consideration as a SARS-CoV-2 antiviral, particularly given its extended use and lack of toxicity in humans, and its availability and affordability,” writes Rocio Martinez-Nunez and colleagues.
“Our data illustrate the power of employing quantum-inspired computing for drug repurposing,” they add.
A pre-print version of the research paper is available on the bioRxiv* server while the article undergoes peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Treatments that can be rapidly deployed are urgently required
Since the COVID-19 outbreak first began in late December 2019, intense research and development efforts have led to the emergency use authorization and mass roll-out of several effective vaccines against SARS-CoV-2.
“However, the ongoing emergence of new variants, different immunization rates, supply chain issues, as well as the presence of smaller or larger outbreaks underlie the requirement for urgent treatments that can be rapidly deployed,” says Martinez-Nunez and colleagues.
Drug repurposing refers to the process by which approved drugs are used to treat a disease they were not initially designed to treat.
Virtual screening has become essential in the early stages of drug discovery, but the process is typically time-consuming since it generally relies on measuring chemical similarities between molecules.
Accordingly, most well-known methods use 2D molecular fingerprints to include structural information about molecules. However, these methods do not consider aspects of molecular structures such as 3D folding.
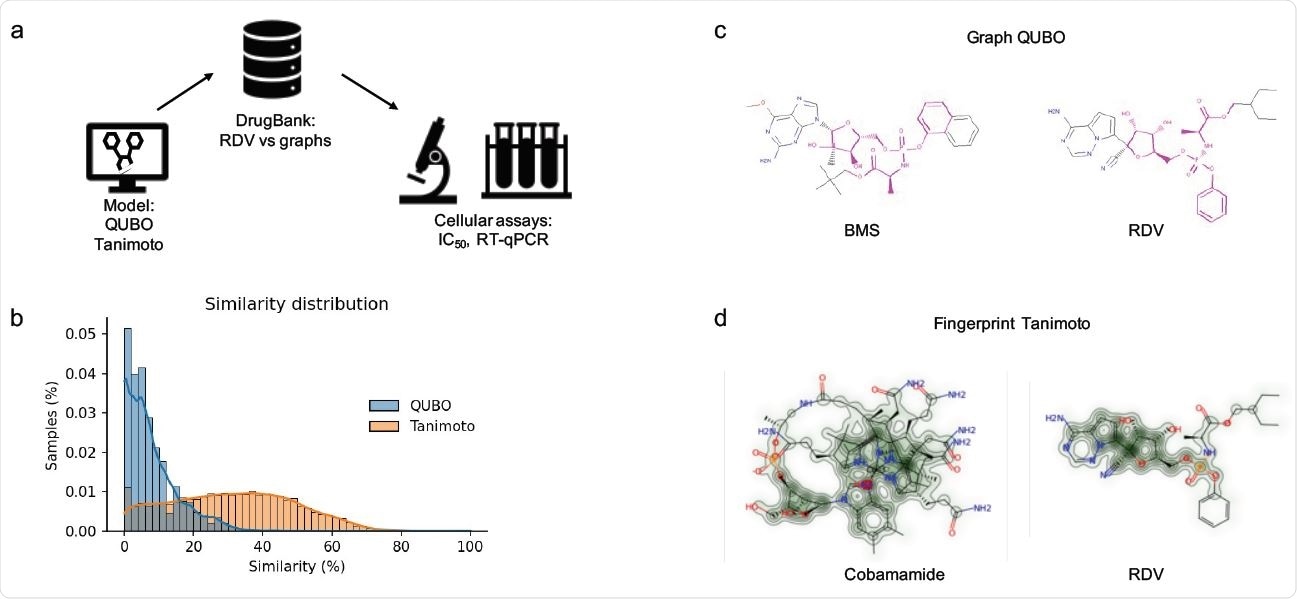
Molecular modeling. (a) pipeline employed in our project. RDV was first modeled as a graph and then screened against the DrugBank dataset. (b) Comparison of samples from the DrugBank predicted as similar to RDV by QUBO (blue) and Tanimoto (orange) models. (c) Graphic representation of BMS-986094 (BMS, left) and RDV (right) similarity according to QUBO. The magenta color represents the similar elements between the two molecules, including atoms as well as bonds, while the rest of the representation is the non-similar elements. d: Graphic representation of similarity for cobamamide (left) and RDV (right) generated by RDkit. The increasing green color represents more similarity between molecules.
Where does quantum-inspired computing come in?
The 3D information obtained from a given molecule can be encoded as a graph, say Martinez-Nunez and colleagues.
“In order to calculate the similarity between molecules, a new graph that contains information regarding the two molecules is required, allowing for better and faster comparisons to solve an optimization problem known as the Maximum Independent Set (MIS) that extracts the similar parts of those two graphs,” explains the team.
By employing quantum-inspired computing, mathematical models are able to manage this kind of information while shortening execution times by up to 60-fold.
The problem with remdesivir
Remdesivir is currently the only antiviral drug approved for use against SARS-CoV-2, while another compound called molnupiravir is also emerging as a potential candidate.
However, both of these compounds are associated with multiple side effects, including nausea and hepatic impairment. They are also costly and therefore unaffordable in many countries and settings.
“There is, therefore, an urgent need to identify novel antiviral compounds that exhibit low to no side effects and that are readily and economically available,” write the researchers.
What did the researchers do?
The researchers employed a Quadratic Unbounded Binary Optimization (QUBO) model that runs on a quantum-inspired device to search for compounds similar to remdesivir.
They modeled remdesivir as a graph and then screened the DrugBank database for compounds already approved for human use. The optimal parameters in the algorithm were established, and the MIS problem was resolved within the conflict graph generated.
The team also employed a more traditional fingerprint method – the Tanimoto index – that runs on a regular laptop.
What did the study find?
Both methods predicted several compounds that exhibited similarity to remdesivir, with GS-6620 predicted as the top compound by both models. The QUBO model predicted BMS-986094 as the second-best candidate and Tanimoto predicted several forms of cobamamide, also known as vitamin B12.
Next, the team performed cultured cell assays to determine the SARS-CoV-2 inhibitory capabilities of the compounds. This revealed that BMS-986094, cobamamide, hydroxocobalamin, and methylcobalamin all proved effective at concentrations lying within ranges suitable for human use.
Finally, the researchers showed that these compounds were effective at inhibiting the replication of a number of SARS-CoV-2 variants, including B.1.1.7 (also called Alpha), B.1.351 (Beta), and B.1.617.2 (Delta).
What did the authors conclude?
Martinez-Nunez and colleagues say the data revealed novel compounds that could inhibit SARS-CoV-2 replication, based on the QUBO model and the more traditional Tanimoto fingerprint.
“BMS warrants further investigation, while vitamin B12 is readily available from multiple sources. It is affordable, can be self-administered by patients, is available worldwide, and displays low-to-no toxicity at high doses,” they write.
“Our screening method can be employed in future searches for novel pharmacologic inhibitors, thus providing an approach for accelerating drug deployment,” concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Rocio T Martinez-Nunez, et al. Drug repurposing based on a Quantum-Inspired method versus classical fingerprinting uncovers potential antivirals against SARS-CoV-2 including vitamin B12. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.06.25.449609, https://www.biorxiv.org/content/10.1101/2021.06.25.449609v1
- Peer reviewed and published scientific report.
Jimenez-Guardeño, Jose M., Ana Maria Ortega-Prieto, Borja Menendez Moreno, Thomas J. A. Maguire, Adam Richardson, Juan Ignacio Diaz-Hernandez, Javier Diez Perez, et al. 2022. “Drug Repurposing Based on a Quantum-Inspired Method versus Classical Fingerprinting Uncovers Potential Antivirals against SARS-CoV-2.” Edited by James M. Briggs. PLOS Computational Biology 18 (7): e1010330. https://doi.org/10.1371/journal.pcbi.1010330. https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1010330.