German experts recently explained why the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) delta variant is more infectious and pathogenic than its ancestor. By conducting a series of in vitro experiments, they have revealed that increased ability to induce cell-to-cell fusion (syncytia) and reduced susceptibility to vaccine and infection-induced antibodies make the delta variant more infectious than previously circulating variants. The study is currently available on the bioRxiv* preprint server.
Since its emergence in late December 2019, SARS-CoV-2, the causative pathogen of coronavirus disease 2019 (COVID-19), has undergone more than 12,000 mutations, the majority of which are neutral, and thus, do not contribute to viral evolution. However, the acquisition of specific mutations in structural and non-structural proteins has caused the emergence of novel, more virulent SARS-CoV-2 variants.
The mutations that appeared in the viral spike protein are particularly vital as they can significantly influence viral infectivity, virulence, and immune evasion ability.
A novel variant of SARS-CoV-2 belonging to the B.1.617 lineage has recently caused a massive surge in new COVID-19 cases in India. The lineage is further divided into three sub-lineages, namely B.1.617.1, B.1.617.2, and B.1.617.3. Although these emerged first in India, the B.1.617.2 variant soon became dominant in many countries, including the United Kingdom.
Because of significantly increased infectivity and pathogenicity, the World Health Organization (WHO) has designated the B.1.617.2 variant as “Variant of Concern” (VOC).
In the current study, the scientists have evaluated the susceptibility of B.1.617.2 variant to neutralization by vaccine or natural infection-induced antibodies.
Mutations in B.1.617.2 variant
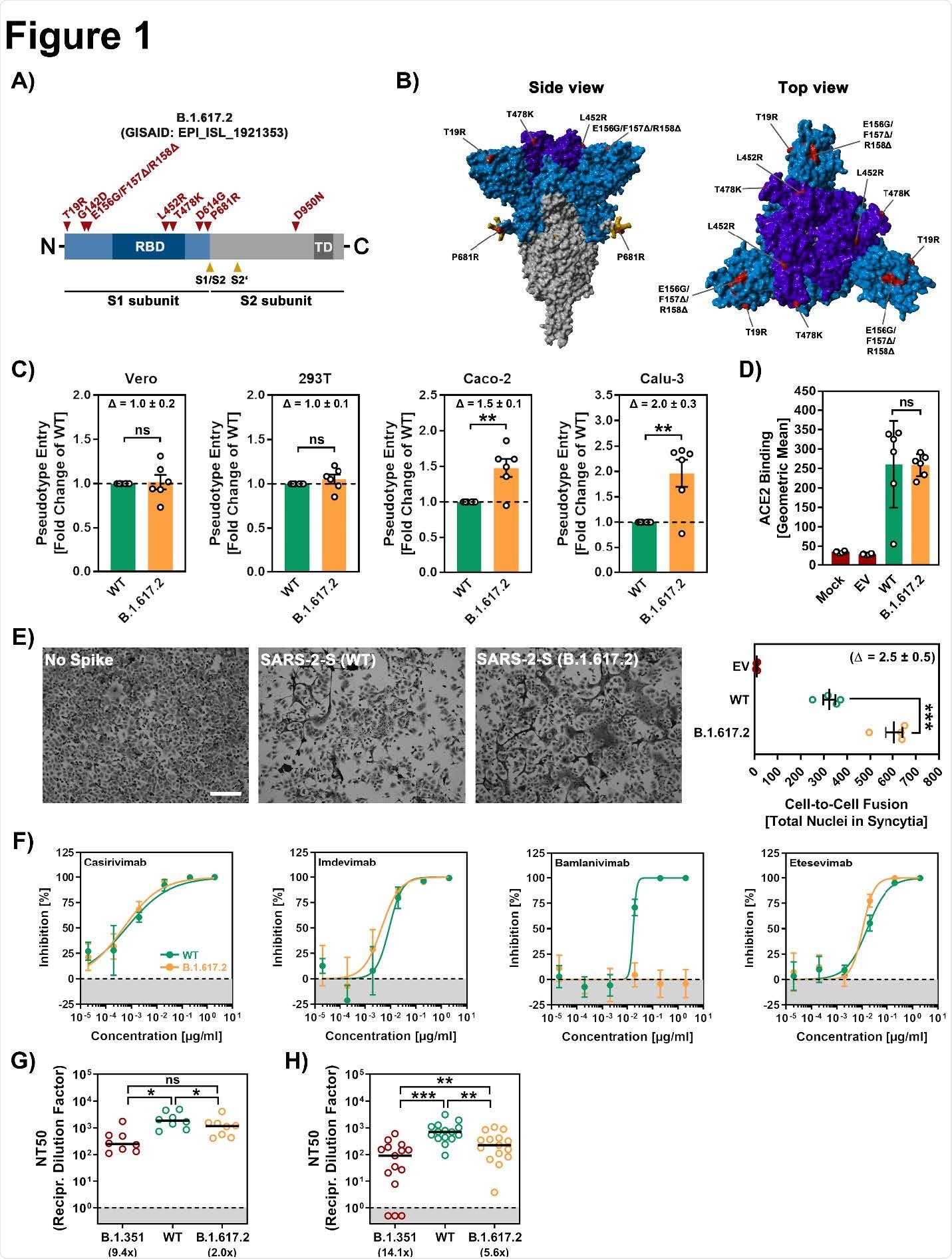
The spike protein of B.1.617.2 variant contains nine mutations in the S1 subunit and one mutation in the S2 subunit. In the S1 subunit, five mutations are present in the N-terminal domain containing binding sites (epitopes) for neutralizing antibodies. In addition, two mutations are present in the receptor-binding domain of the S1 subunit, which is known to influence antibody-mediated neutralization and infectivity. Among the three remaining mutations, two are known to increase angiotensin-converting enzyme 2 (ACE2) binding, viral replication, and spike protein cleavage at the S1/S2 site.
Host cell entry of B.1.617.2 variant
To investigate whether B.1.617.2 has a higher ability to enter host cells, the scientists conducted a series of experiments on African green monkey kidney cells and human kidney, colon, and lung cells that express endogenous ACE2.
The findings revealed that B.1.617.2 can enter human and monkey kidney cells with similar efficacy as the wild-type SARS-CoV-2. However, for human colon and lung cells, B.1.617.2 showed 1.5-fold and 2-fold higher invading ability, respectively, compared to the wild-type virus.
These observations indicate that spike protein-mediated host cell entry may vary between cell types and that B.1.617.2 has higher efficacy in entering lung and colon cells.
Since the spike protein of B.1.617.2 did not exhibit increased ACE2 binding, the scientists suggest that increased entry of B.1.617.2 into colon and lung cells is not mediated by enhanced ACE2 binding.
Besides inducing viral envelop – host cell membrane fusion, the spike protein triggers the fusion of infected cells with nearby cells to form large multinucleated cells (syncytia). Given the fact that spike-induced syncytia formation contributes to COVID-19 pathogenesis, the scientists investigated whether B.1.617.2 infection is associated with increased syncytia formation.
By conducting in vitro experiments on human lung cells expressing high levels of ACE2, they revealed that expression of wild-type spike results in syncytia formation, whereas expression of B.1.617.2 spike leads to 2.5-fold higher and larger syncytia formation.
Cell entry and evasion of antibody-mediated neutralization by the spike protein of SARS-CoV-2 B.1.617.2. (A) Transduction data normalized against the assay background (related to Figure 1C). The experiment was performed as described in the legend of Figure 1C. Presented are the average (mean) data from the same six biological replicates (each conducted with technical quadruplicates) as presented in Figure 1C with the difference that transduction was normalized against signals obtained from cells inoculated with particles bearing no viral glycoprotein (background, set as 1). In addition, transduction data of particles bearing VSV-G are included. Error bars indicate the SEM. (B) Location of the receptor binding domain (gray) mutations L452R and T478K (both red) of SARS-CoV-2 variant B.1.617.2 in the context of the interfaces for ACE2 binding (orange) and binding of monoclonal antibodies used for COVID-19 therapy. (C) An unrelated control antibody does not affect cell entry of pseudotype particles bearing SARS-CoV-2 WT, B.1.351 or B.1.617.2 S (D) Individual neutralization data for convalescent plasma (related to Figure 1F). Pseudotype particles bearing the indicated S proteins were incubated (30 min, 37 °C) with different dilutions of convalescent plasma before being inoculated onto Vero cells. Transduction efficiency was quantified by measuring virus-encoded luciferase activity in cell lysates at 16-18 h posttransduction. Presented are the data from a single representative experiment conducted with technical quadruplicates. For normalization, inhibition of S protein-driven entry in samples without plasma was set as 0%. Error bars indicate the SD. The data were further used to calculated the plasma/serum dilution that leads to 50% reduction in S protein-driven cell entry. (E) Individual neutralization data for vaccinee serum (related to Figure 1G). Pseudotype particles bearing the indicated S proteins were incubated (30 min, 37 °C) with different dilutions of serum from individuals vaccinated with the Pfizer/BioNTech vaccine Comirnaty/BNT162b2 before being inoculated onto Vero cells. Transduction efficiency was quantified by measuring virus31 encoded luciferase activity in cell lysates at 16-18 h posttransduction. Presented are the data from a single representative experiment conducted with technical quadruplicates. For normalization, inhibition of S protein-driven entry in samples without plasma was set as 0%. Error bars indicate the SD. The data were further used to calculated the NT50 shown.
Immune evasion ability of B.1.617.2 variant
The scientists tested the efficacy of four therapeutic monoclonal antibodies in neutralizing the B.1.617.2 variant. Of tested antibodies, only Bamlanivimab failed to neutralize B.1.617.2. The other three antibodies exhibited similar efficacy in neutralizing both wild-type virus and B.1.617.2 variant. These observations indicate that Bamlanivimab monotherapy may not be effective in treating B.1.617.2 infection.
Significantly, antibodies derived from COVID-19 recovered patients, and BNT162b2-vaccinated individuals showed slightly reduced efficacy in neutralizing the B.1.617.2 variant as compared to the wild-type virus. In contrast, the B.1.315 variant, which was first detected in South Africa, showed a significantly higher ability to evade infection- and vaccination-induced immunity.
Study significance
The study reveals that the increased ability of the B.1.617.2 variant to invade lung cells may enhance infectivity and pathogenicity. Despite lower susceptibility to antibody-mediated neutralization, B.1.617.2 can be effectively controlled by immunity developed in response to natural infection or vaccination.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources