There has been an unprecedented research effort to understand the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Despite this, the subcellular distribution of coronavirus in COVID-19 patients' lungs remains unknown.
Scientists from the Netherlands and the US recently studied the localization of the SARS-CoV-2 proteins in postmortem lung tissues of COVID-19 patients and in SARS-CoV-2 infected Vero cells that were processed identically. The research is posted to the bioRxiv* preprint server, prior to peer review.
Visualizing viral particles in SARS-CoV-2-infected cells and detecting viral proteins and the ultrastructure
The researchers used correlative light and electron microscopy on semi-thick cryosections to demonstrate the induction of electron-lucent, lipid-filled subcellular compartments post SARS-CoV-2 infection in both cell and lung cultures. The objective of this study was to visualize virus particles in SARS-CoV-2-infected cells by focusing on the detection of viral protein in combination with the ultrastructure.
With the help of immuno-electron microscopy, viral proteins were detected in these lipid-filled compartments in infected Vero cells. Additionally, they also detected many viral proteins in Golgi, virus particles, double-membrane spherules, and non-lysosomal multiple-virus bodies. The non-structural protein 4 and the stable nucleocapsid N- protein were detected in the membranes of lipid-containing compartments in the lung tissue.
Accumulation of lipids and viral proteins in the lung of severe COVID-19 patients may explain the severe inflammatory response
According to the authors, the localization of the viral proteins and the induction of lipid-filled compartments in the lung of patients with severe COVID-19 may explain the severe inflammatory response in these patients. They analyzed how the virus altered cell morphology and showed that lipid-filled compartments in Vero cells contained many different viral proteins.
.jpg)
Lipid accumulates in e-lucent compartments more densely in infected Vero cells. Fluorescence microscopy of DNA and lipid staining with Nile red in A) the uninfected control (Con) Vero cells and B) cells infected with SARS-CoV-2 for 24 hours; C) Electron microscopy of infected cells; D) Fluorescence microscopy of the same cells, and E) Correlative light-electron microscopy (CLEM) showing lipid staining at e-lucent compartments in the electron microscope. Immuno-EM labeling for lipid droplet marker perilipin-2 in F) uninfected Vero cells and G) cells infected with SARS-CoV-2 for 24 hours. Blue color in A, B, D, and E shows the nuclei stained with Hoechst and red shows the lipids stained with Nile red. In electron micrographs, lipid like structure is denoted by red arrows, virus particles by black arrows, immuno-gold labeling of perilipin-2 by red circles, mitochondria by m, and lipid droplets by LD.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
In the Vero cells, they also found membrane-enclosed multi-virus bodies with a different set of viral proteins. This demonstrated that lipid-filled compartments are viral-induced compartments since there were no known cellular markers like lipid droplets or lysosomal markers in the compartments.
"We demonstrated that lipid-filled compartments are viral induced compartments, as no known cellular marker such as lipid droplet or lysosomal marker was present."
With the help of this finding, they then analyzed similarly processed lung tissue from fatal COVID-19 patients. Once again, they detected lipid accumulation in the compartments, but now these compartments had viral proteins nsp4 and the stable SARS-CoV-2 nucleocapsid N-protein. Thus, they concluded that these lipid-filled compartments with SARS proteins induced by viral infection might be the reason the immune response of the patients with severe COVID-19 is so intense, resulting in fatal reactions.
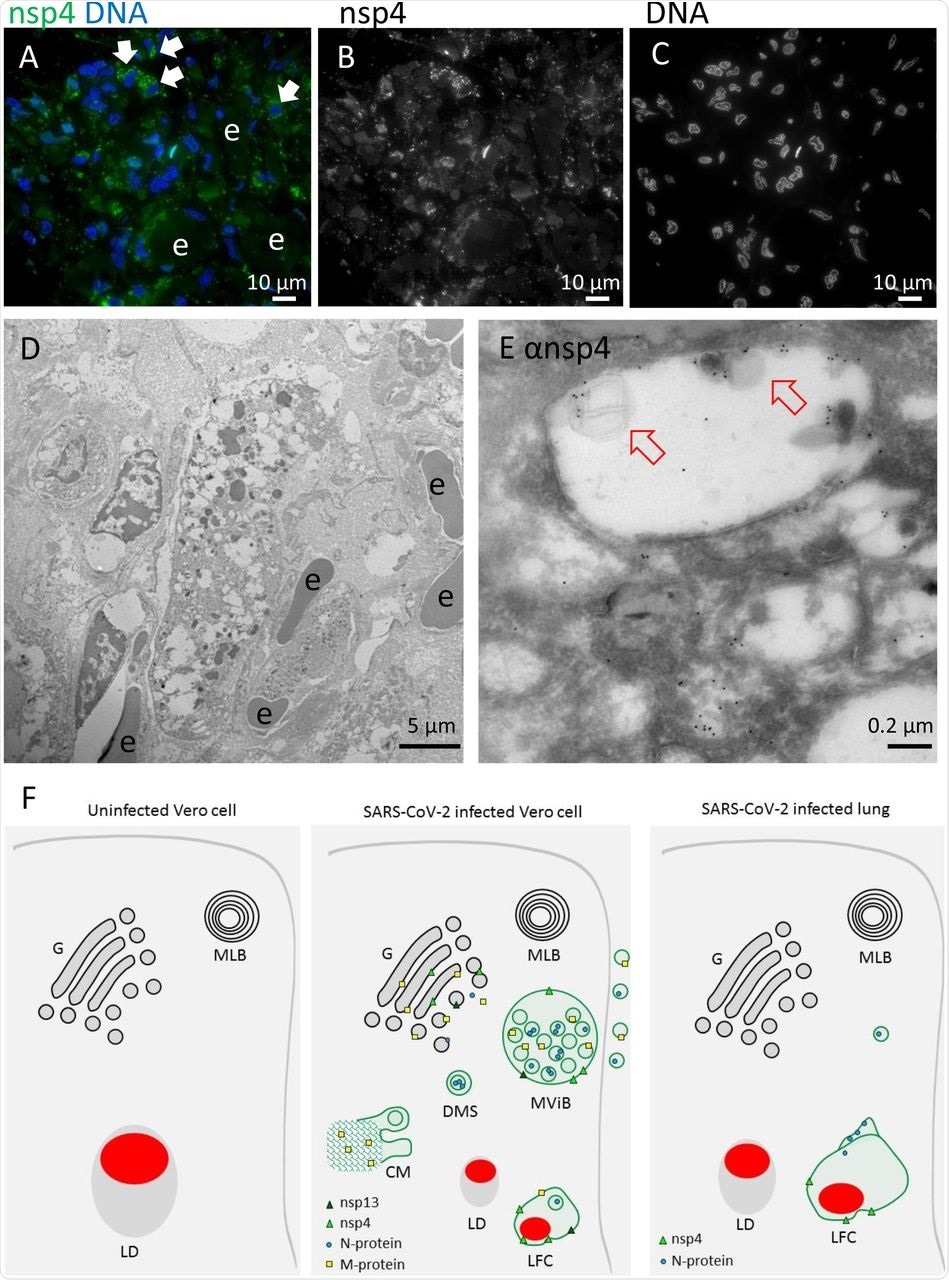
Non-structural protein 4 in e-lucent compartments infected lung. Lung tissue of COVID-19 patient 58 was either sectioned semi-thin for FM with A) nuclei, stained with Hoechst (blue), nsp4 stained with Alexa (green) in nsp4 positive cells indicated by white arrows and in black and white, and erythrocytes represented by e. Separate channels of nsp4 (B) and DNA (C). Ultrathin sections of infected lung immuno-gold labeled against nsp-4 and 10-nm gold particles in overview (D) and at higher magnification (E) e-lucent compartments with nsp4 labeling on membrane and lipid-like structures (open red arrows) erythrocytes represented by e. F) Schematic representation of uninfected Vero cells, SARS-CoV-2 infected Vero cells and lung tissue of COVID-19 patient summarizing presence of cellular organelles and subcellular localization viral proteins. In black: host compartments, in green: viral compartments, in red: lipid-like structures, CM convoluted membrane DMS: double membrane spherules, G: Golgi, LD: lipid droplet, MLB: multi-lamellar bodies, MViB: multi-virus body, LFC: lipid-filled compartment and immuno-labeling viral proteins: dark green triangle: nsp13, light green triangle: nsp4, blue circle: N-protein, yellow square: M-protein.
Findings of the study could help develop new COVID-19 treatment strategies
To summarize, viral particles are difficult to detect in the lungs even after a fatal SARS-CoV-2 infection, but immuno-EM showed that SARS-CoV-2 virus particles are 90-nm spherical and 110-nm oval particles in Vero cells. Non-structural proteins nsp4 and 13 were detected in electron-lucent and partly lipid-filled compartments induced by SARS-CoV-2 infection in Vero cells. In lung tissues, similar lipid accumulation was detected in the sites of N-protein and nsp4 accumulation.
"The induction of such lipid-filled compartments and the localization of the viral proteins in the lung of patients with fatal COVID-19 might explain the extensive inflammatory response."
Thus, the authors speculate that lipid-filled compartments with viral proteins may have an important role in the secondary effects of the COVID-19. The uncontrolled immune responses, including the cytokine storm that causes severe damage in fatal COVID-19, might be a response to either the proteins or the lipids accumulating in these subcellular compartments. This finding can help modify new therapeutic strategies for COVID-19.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Lipid and nucleocapsid N-protein accumulation in COVID-19 patient lung and infected cells Anita E. Grootemaat, Sanne van der Niet, Edwin R. Scholl, Eva Roos, Bernadette Schurink, Marianna Bugiani, Sara E. Miller, Per Larsen, Jeannette Pankras, Eric A. Reits, Nicole N. van der Wel bioRxiv 2021.06.24.449252; doi: https://doi.org/10.1101/2021.06.24.449252, https://www.biorxiv.org/content/10.1101/2021.06.24.449252v1
- Peer reviewed and published scientific report.
Grootemaat, Anita E., Sanne van der Niet, Edwin R. Scholl, Eva Roos, Bernadette Schurink, Marianna Bugiani, Sara E. Miller, et al. 2022. “Lipid and Nucleocapsid N-Protein Accumulation in COVID-19 Patient Lung and Infected Cells.” Edited by Benjamin W. Neuman. Microbiology Spectrum 10 (1). https://doi.org/10.1128/spectrum.01271-21. https://journals.asm.org/doi/10.1128/spectrum.01271-21.