Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes various symptoms that go beyond respiratory problems. Neurological, cardiac, and other issues can emerge during infection and after recovery in long COVID, suggesting SARS-CoV-2 invades several organs. However, how SARS-CoV-2 invades the brain — whether directly or indirectly — remains under investigation.
New research led by Le Cong of Stanford University School of Medicine in California indicates that the KCNA6 potassium channel may help with viral entry and promote infection. In addition, KCNA6 is highly expressed in OLIG2+ neuronal cells, which has been shown to be suspectable to SARS-CoV-2 in the olfactory neuroepithelium. This area appears to contribute to loss of smell commonly observed during infection.
The study authors write:
“We have thus identified the potassium channel KCNA6 as a SARS-CoV-2 host factor, expanded our understanding of potential viral tropism, and identified promising targets for drug repurposing and development.”
The study “CRISPRa screening with real-world evidence identifies potassium channels as neuronal entry factors and druggable targets for SARS-CoV-2” is available as a preprint on the bioRxiv* server, while awaiting formal peer review.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
How they did it
The researchers used CRISPR activation (CRISPRa) screening to look at the potential viral entry into the body. Because SARS-CoV-2 affects organs beyond the lungs, they first looked for specific host factors that would make a cell type susceptible to infection.
Looking at a virus’s ability to infect different cell types may require multiple analyses of multiple cell lines representing various cell types, but this requires an arduous amount of time and prior knowledge of viral tropism.
The researchers used CRISPRa to circumvent these issues by “selecting cell lines with limited or no susceptibility and allowing for the unbiased determination of factors that promote viral entry.”
CRISPRa screened and targeted 6,213 human membrane proteins with approximately 24,00 sgRNAs with non-targeting controls. They included membrane proteins that may sometimes be excluded in genome libraries but can hide hidden host factors.
A pseudo-typed lentivirus expressing the SARS-CoV-2 spike protein or vesicular stomatitis virus envelope G protein as a control was used to observe viral infection.
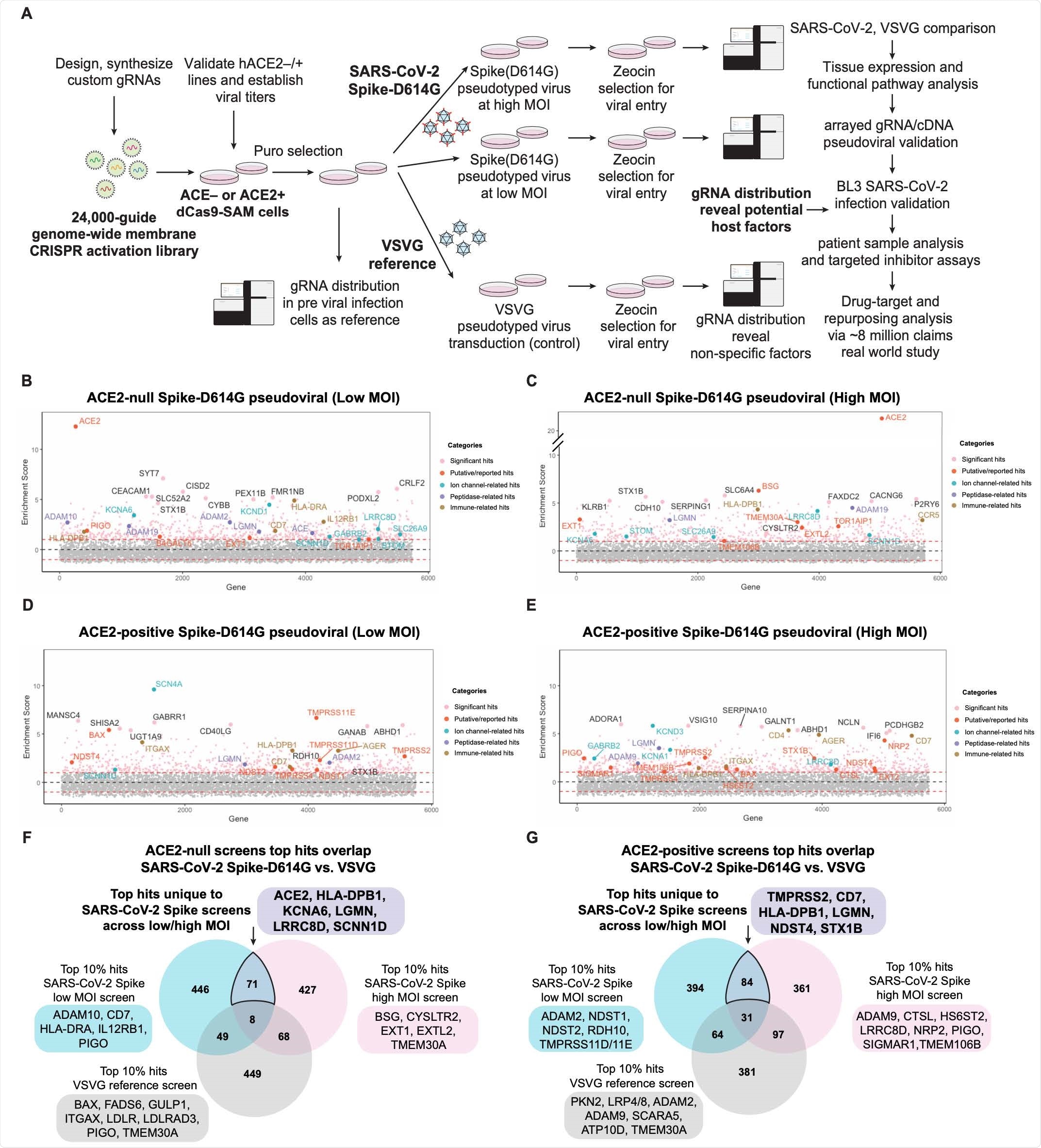
Membrane-focused CRISPRa screening identifies potential host factors involved in Spike-dependent SARS-CoV-2 virus entry. (A) Screen pipeline showing different conditions used (ACE2-null, ACE2-positive, at low or high MOI, with VSVG references), downstream analyses and validation workflow. (B-E) Enrichment scores of CRISPRa screen across different conditions with top hits highlighted and colored by their functional categories. (F-G) Differential analysis of top 10% hits from SARS-CoV-2 Spike and reference VSVG screens. The unique hits in SARS-CoV-2 screens identify putative virus-specific host factors.
KCNA6 potassium channel is a target for SARS-CoV-2 infection
CRISPRa screening showed previously unknown host factors in several areas of the body that can increase viral entry. This includes neuronal (KCNA6), immune (HLADPB1, CD7), and cardiac (LGMN, EPHA4).
Specifically, the team found the voltage-gated potassium channel known as KCNA6 and found on neuronal cells is susceptible to SARS-CoV-2 entry and infection. Viral entry was observed even when ACE2 expression levels were low or undetectable. Furthermore, overexpressing the KCNA6 channel increased the likelihood of infection among the spike protein of the Beta (B.1.351) SARS-CoV-2 variant.
Having low ACE2 expression does not appear to affect SARS-CoV-2 infection as both B.1.351, and the D614G variant were capable of infecting cells overexpressing KCNA6.
“This could be significant in light of reports that emerging SARS-CoV-2 variants are often less dependent on ACE2 binding and resistant to antibody therapeutics,” concluded the team.
High KCNA6 expression with low ACE2 was identified on OLIG2+ cells, which are found in the human olfactory neuroepithelium. The results suggest KCNA6 may play a role in the loss of smell and other neuronal symptoms reported during COVID-19 infection.
Potassium channel blockers as potential coronavirus treatment
KCNA6 may be a future therapeutic target worth exploring. Looking at about 8 million patient records in insurance claims showed an association of loop diuretics, SSRI antidepressants, and broad-spectrum anticonvulsants that target potassium channels and less severe COVID-19 illness not requiring hospitalization.
The results suggest prescription drugs could be repurposed as treatments for COVID-19.
In addition, the study results showed that applying the KCNA channel blocker 4-AP (dalfampridine) reduced viral entry in a dose-dependent manner. It also suggests SARS-COV-2 may potentially be affecting KCNA potassium channels more generally rather than only KCNA6.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.