A new paper discusses an intriguing new mechanism of immune evasion traceable to specific mutations in the B.1.427/B.1.429 variant of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
The coronavirus disease 2019 (COVID-19), which is caused by SARS-CoV-2, causes a wide range of symptoms from asymptomatic cases to acute respiratory distress syndrome (ARDS). Patients with severe cases of COVID-19 can experience extra-pulmonary complications including clotting disorders and the cytokine storm. Both of these complications can contribute to multi-system dysfunction, which subsequently increases the likelihood of death.
SARS-CoV-2 infects the host cell via its spike (S) glycoprotein, which binds to the angiotensin-converting enzyme 2 (ACE2) entry receptor on host cells. The S protein consists of two subunits referred to as S1 and S2 subunits.
The S1 subunit contains the receptor-binding domain (RBD) of the S protein, which directly interacts with the ACE2 receptor on host cells. More specifically, the RBD interacts with the ACE2 receptor via residues on the receptor-binding motif (RBM).
The S1 subunit also contains the N-terminal domain (NTD), which binds with the DC-SIGN, L-SIGN, and AXL receptors. This binding activity may serve to attach the virus to the host cell. In both vaccine recipients and individuals who have recovered from COVID-19, neutralizing antibodies target both the RBD and the NTD.
Newer variants
New emerging variants of SARS-CoV-2 are constantly being detected, such as the D614G variant that rapidly rose to dominance worldwide. Currently, the B.1.1.7, B.1.351, and P.1 lineages are well-known, all of which contain a number of S protein mutations, among others. Several of these mutations are linked to reduced neutralization by monoclonal antibodies (mAbs), convalescent sera, as well as neutralizing antibodies elicited by the messenger ribonucleic acid (mRNA) vaccines (Pfizer/BioNTech and Moderna).
Currently, the B.1.1.7 variant is one of the prominent strains of SARS-CoV-2 that is currently circulating worldwide as a result of its increased transmissibility. This change underlines the need to monitor the occurrence of antigenic drift in SARS-CoV-2.
The B.1.427/B.1.429 lineage has S protein mutations and L452R in the RBD, as well as S13I, W152C in the NTD, along with mutations in other regions of the SARS-CoV-2 genome. Both are thought to have emerged in between June and July 2020. Until April 30, 2021, over 8,400 and 21,000 genomes of these strains have been sequenced, respectively, from the United States, as well as more than 30 other countries worldwide.
From February 2021, over half the sequenced genomes from California belonged to one of these two strains. This rapid spread over California, the rest of the United States, and other countries indicate the increased transmissibility of this virus relative to the wild-type virus.
Reduced neutralization potency
The scientists of the current study carried out direct comparisons of neutralizing capacity with plasma from individuals who had received two doses of either of the mRNA vaccines, from a week to a month after the second dose. All samples contained neutralizing antibodies; however, the average potency of neutralization was reduced by approximately two-fold when tested against pseudoviruses carrying B.1.427/B.1.429 S protein.
When plasma from individuals who had been infected with COVID-19 prior to receiving two doses of the vaccine was tested, the neutralization potency was diminished by approximately three-fold against B.1.427/B.1.429 and B.1.351 S protein, compared to D614. When this same plasma sample was tested against the B.1.1.7 and P.1 variants, neutralizing potency was reduced by 1.3- and 1.7-fold.
Convalescent plasma from individuals with a history of symptomatic COVID-19 who had been infected early on in the pandemic, before the D614G strain became dominant, was also studied. When these plasma samples were tested, the neutralization potency was reduced by approximately 3.3-fold for B.1.427/B.1.429, and for P.1, and by 4.4-fold for B.1.351. However, the B.1.1.7 strain was less resistant, with the neutralizing activity falling by approximately two-fold.
Thus, the three S protein mutations were found to significantly alter the neutralization potency of both vaccine- and infection-elicited antibodies. However, antibodies elicited by natural infection were more sensitive to these mutations as compared to those induced by vaccination.
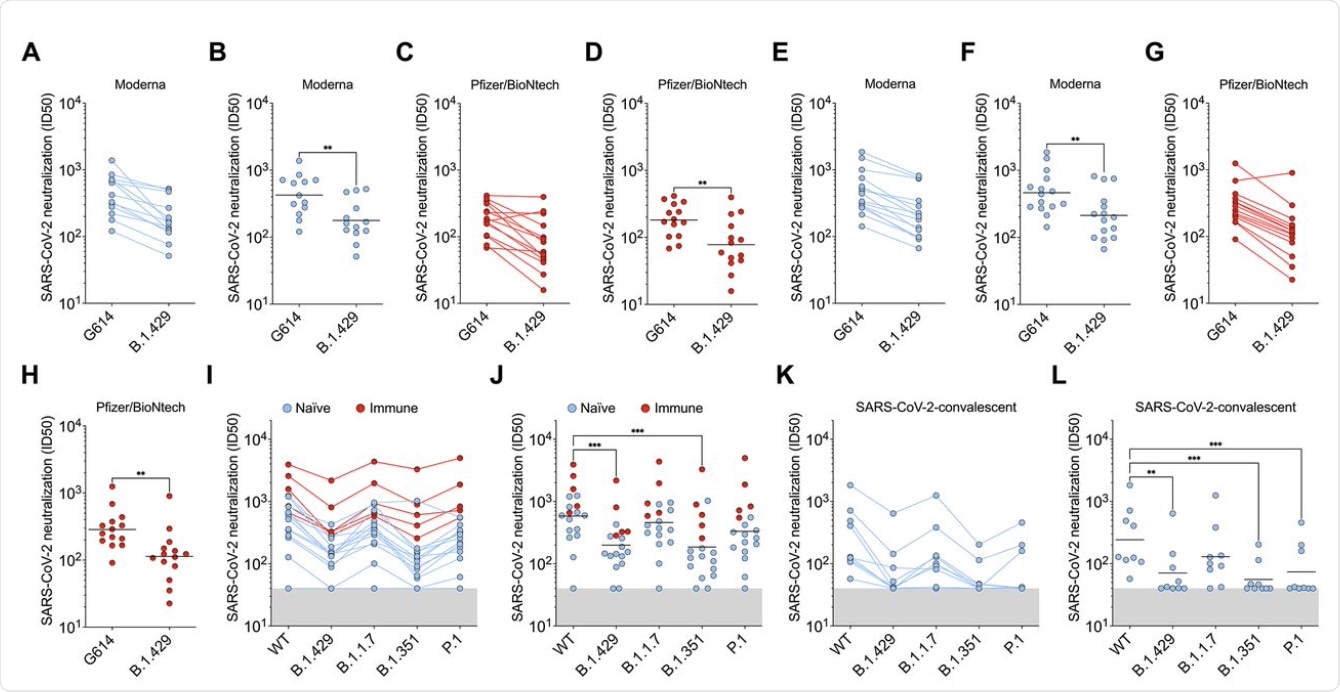 B.1.427/B.1.429 (B.1.429) S determined using plasma from individuals who received two doses of Pfizer/BioNtech BNT162b2 mRNA vaccine (red). (I and J) Neutralizing Ab ID50 (I) and GMT (J) titers against VSV pseudotyped viruses harboring D614 SARS-CoV-2 S, B.1.427/B.1.429 S, B.1.1.7 S, B.1.351 S, or P.1 S determined using plasma from naïve (blue) and previously infected (red) individuals who received two doses of Pfizer/BioNtech BNT162b2 mRNA vaccine. Naïve: vaccinated individuals who had not been previously infected with SARS-CoV-2. Immune: vaccinated individuals who had been previously infected with SARS-CoV-2. (K and L) Neutralizing Ab ID50 (K) and GMT (L) titers against VSV pseudotyped viruses harboring D614 SARS-CoV-2 S, B.1.427/B.1.429 S, B.1.1.7 S, B.1.351 S or P.1 S determined using plasma from convalescent individuals who were infected with wildtype SARS-CoV-2. Neutralization data shown in (A) to (H) and (I) to (L) were performed using 293T-ACE2 and VeroE6-TMPRSS2, respectively. Data are average of n = 2 replicates.
B.1.427/B.1.429 (B.1.429) S determined using plasma from individuals who received two doses of Pfizer/BioNtech BNT162b2 mRNA vaccine (red). (I and J) Neutralizing Ab ID50 (I) and GMT (J) titers against VSV pseudotyped viruses harboring D614 SARS-CoV-2 S, B.1.427/B.1.429 S, B.1.1.7 S, B.1.351 S, or P.1 S determined using plasma from naïve (blue) and previously infected (red) individuals who received two doses of Pfizer/BioNtech BNT162b2 mRNA vaccine. Naïve: vaccinated individuals who had not been previously infected with SARS-CoV-2. Immune: vaccinated individuals who had been previously infected with SARS-CoV-2. (K and L) Neutralizing Ab ID50 (K) and GMT (L) titers against VSV pseudotyped viruses harboring D614 SARS-CoV-2 S, B.1.427/B.1.429 S, B.1.1.7 S, B.1.351 S or P.1 S determined using plasma from convalescent individuals who were infected with wildtype SARS-CoV-2. Neutralization data shown in (A) to (H) and (I) to (L) were performed using 293T-ACE2 and VeroE6-TMPRSS2, respectively. Data are average of n = 2 replicates.
Monoclonal antibodies show reduced neutralizing efficacy
Among the panel of 34 mAbs directed against the RBD, including six that are currently in clinical use, 14 showed a reduction in neutralizing potency against B.1.427/B.1.429 as compared to D614. Of the six clinical mAbs, bamlanivimab (LY-CoV555) was found to lose its neutralizing activity completely, while two others (etesevimab (LY-CoV016) and, more significantly, regdanvimab (CT-P59), showed reduced potency.
The cocktail of Regeneron antibodies (REGN10933 and REGN10987) or casirivimab/imdevimab, as well as sotrovimab , which is another type of monoclonal antibody, were all found to retain potency against each of these SARS-CoV-2 variants.
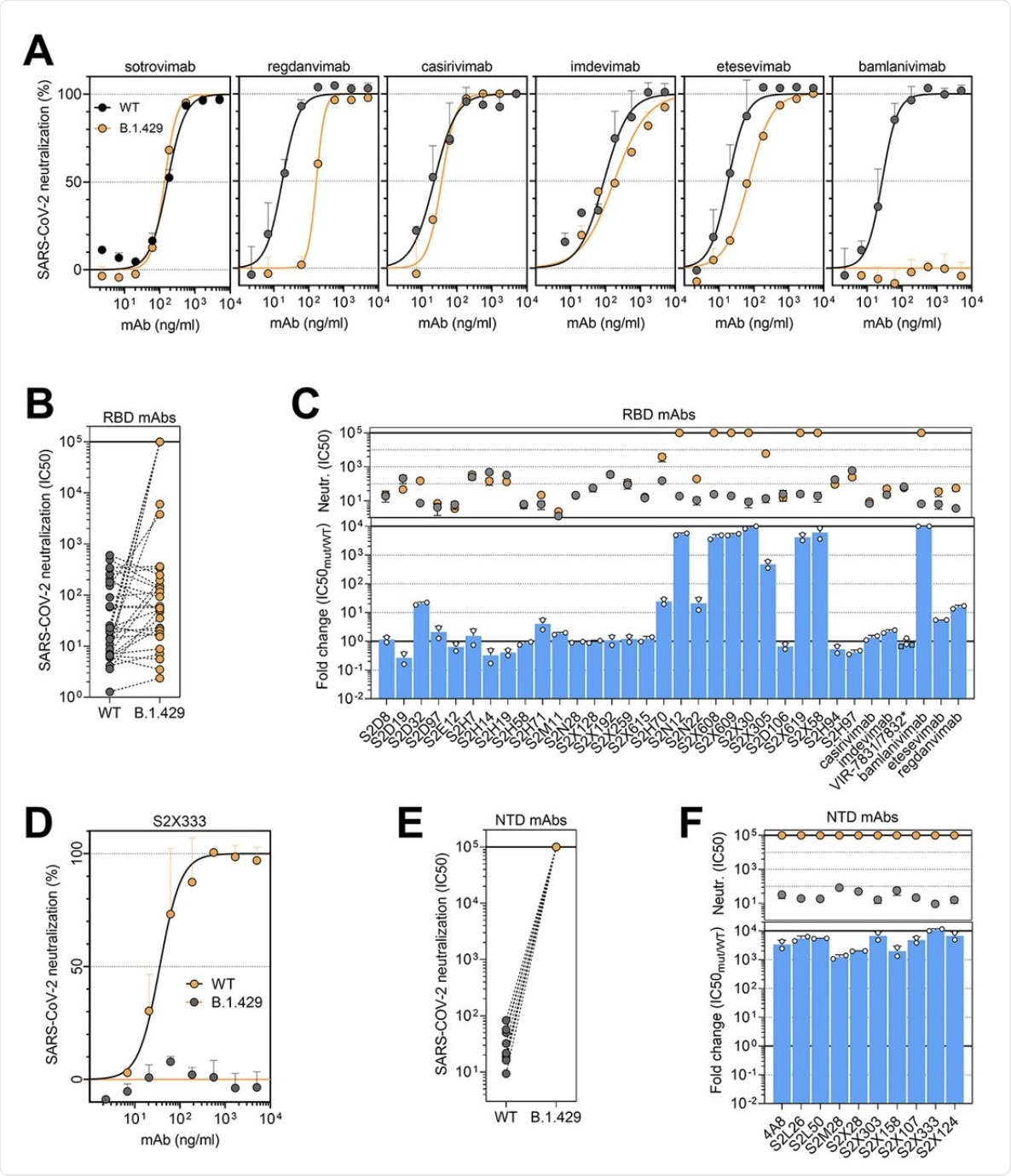 (A and D) Neutralization of SARS-CoV-2 pseudotyped VSV carrying D614 (grey) or B.1.427/B.1.429 (orange) S protein by clinical-stage RBD mAbs (A) and an NTD-targeting mAb (S2X333) (D). Data are representative of n = 2 replicates. (B and E) Neutralization of SARS-CoV-2 S VSV pseudotypes carrying D614 or B.1.427/B.1.429 S by 34 mAbs targeting the RBD and 10 mAbs targeting the NTD. Data are the mean of 50% inhibitory concentration (IC50) values (ng/ml) of n = 2 independent experiments. Non-neutralizing IC50 titers were set at 105 ng/ml. (C and F) Neutralization by RBD-specific (C) and NTD-specific (G) mAbs showed as mean IC50 values (top) and mean fold change (bottom) for B.1.427/B.1.429 S (orange) relative to D614G S (grey) VSV pseudoviruses. VIR-7831 is a derivative of S309 mAb (sotrovimab). *, VIR-7832 (variant of VIR-7831 carrying the LS-GAALIE Fc mutations) shown as squares. Non-neutralizing IC50 titers and fold change were set to 105 ng/ml and 104, respectively.
(A and D) Neutralization of SARS-CoV-2 pseudotyped VSV carrying D614 (grey) or B.1.427/B.1.429 (orange) S protein by clinical-stage RBD mAbs (A) and an NTD-targeting mAb (S2X333) (D). Data are representative of n = 2 replicates. (B and E) Neutralization of SARS-CoV-2 S VSV pseudotypes carrying D614 or B.1.427/B.1.429 S by 34 mAbs targeting the RBD and 10 mAbs targeting the NTD. Data are the mean of 50% inhibitory concentration (IC50) values (ng/ml) of n = 2 independent experiments. Non-neutralizing IC50 titers were set at 105 ng/ml. (C and F) Neutralization by RBD-specific (C) and NTD-specific (G) mAbs showed as mean IC50 values (top) and mean fold change (bottom) for B.1.427/B.1.429 S (orange) relative to D614G S (grey) VSV pseudoviruses. VIR-7831 is a derivative of S309 mAb (sotrovimab). *, VIR-7832 (variant of VIR-7831 carrying the LS-GAALIE Fc mutations) shown as squares. Non-neutralizing IC50 titers and fold change were set to 105 ng/ml and 104, respectively.
The researchers established that the loss of immune neutralization was due to the L452R mutation. This conclusion was due to the fact that all ten mAbs specific to the RBD showed a ten-fold or more loss of potency against the B.1.427/B.1.429 variant compared to D614. Furthermore, these ten mAbs were found to show low binding affinity to the S protein bearing this mutation in the RBD.
The non-RBD S protein and NTD mutations of S13I and W152C led to complete loss of neutralizing activity of all ten mAbs that targeted the NTD. Thus, immune evasion can result from the evasion of neutralizing antibodies directed against both these regions.
Signal peptide modification leads to immune evasion
The loss of neutralizing capacity by regdanvimab and bamlanivimab was explained by structural studies that showed steric hindrance by the L452R mutation, thus making it difficult for these antibodies to bind to the RBD mutant. At this residue, the B.1.427/B.1.429 mutations are directed away from the ACE2 receptor. Furthermore, these mutations not take part in the S-receptor engagement, thereby suggesting that they are not involved in disrupting this binding.
Instead, both the B.1.427/B.1.429 variant and wildtype RBD demonstrated equivalent binding affinities for ACE2.
The NTD antigenic supersite of the B.1.427/B.1.429 variants underwent marked alteration. These changes account for the loss of binding and neutralization by the array of anti-NTD antibodies, both mAbs, as well as those detected in vaccine-induced plasma.
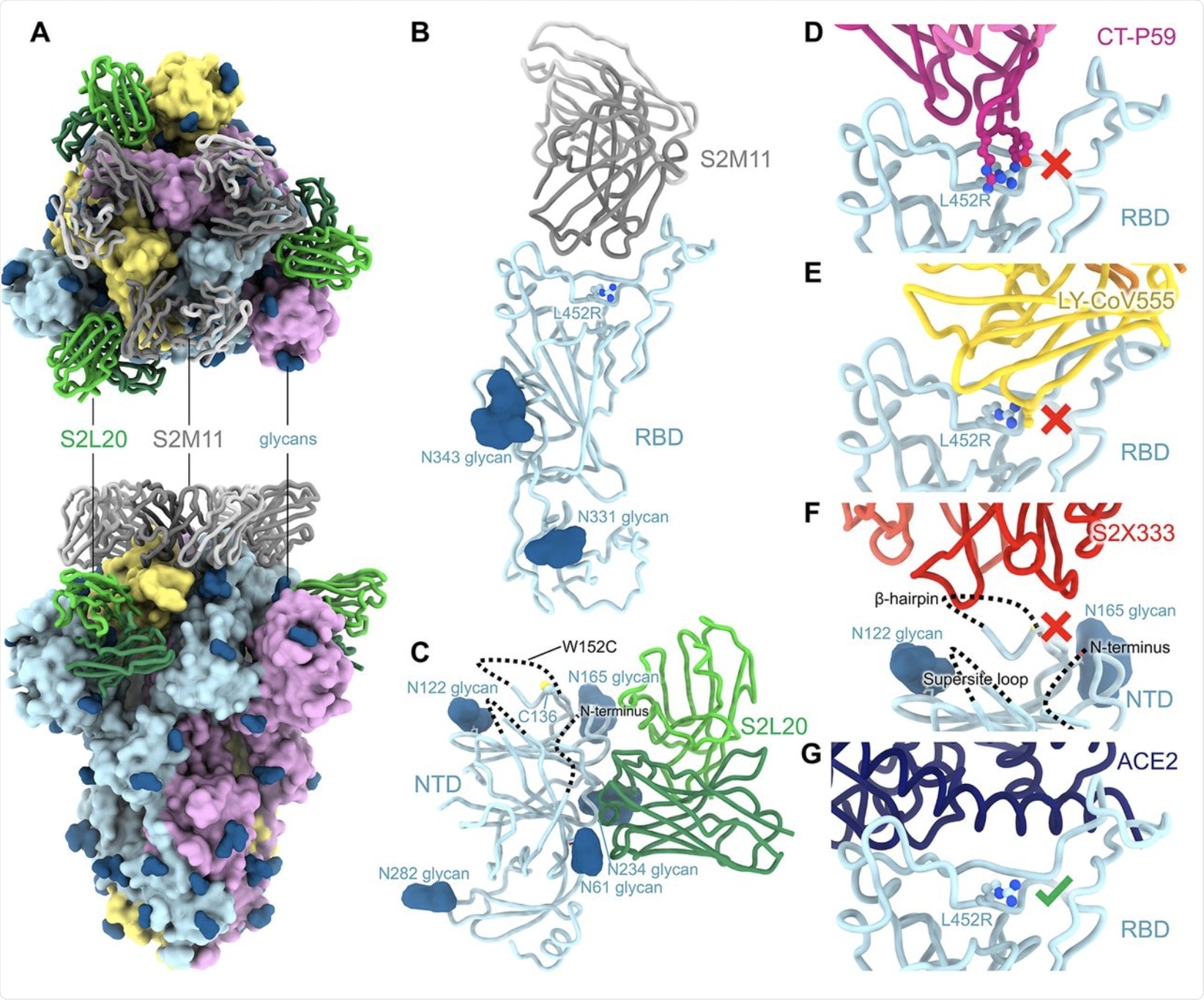 CryoEM structure of the SARS-CoV-2 B.1.427/B.1.429 S ectodomain trimer. (A) Structure of the S trimer (surface rendering) bound to the S2M11 and S2L20 Fabs (ribbons) in two orthogonal orientations. SARS-CoV-2 S protomers are colored pink, cyan, and gold, whereas the S2L20 Fab heavy and light chains are colored dark and light green, respectively, and the S2M11 Fab heavy and light chains are colored dark and light gray, respectively. Only the Fab variable domains are resolved in the map. N-linked glycans are rendered as dark blue spheres. (B) Zoomed in view of the S2M11-bound RBD with R452 shown in ball and stick representation. (C) Zoomed in view of the S2L20-bound NTD with disordered N terminus, supersite β-hairpin and loop regions shown as dashed lines. (D) Superimposition of the CT-P59–bound SARS-CoV-2 RBD structure (PDB 7CM4) on the SARS-CoV-2 B.1.427/B.1.429 S cryoEM structure show that R452 would sterically clash with the mAb. (E) Superimposition of the LY-CoV555–bound SARS-CoV-2 RBD structure (PDB 7KMG) on the SARS-CoV-2 B.1.427/B.1.429 S cryoEM structure show that L452R would sterically clash with the mAb. (F) Superimposition of the S2X333-bound SARS-CoV-2 S structure (PDB 7LXW) on the SARS-CoV-2 B.1.427/B.1.429 S cryoEM structure reveals that most of the NTD antigenic supersite epitope residues are disordered. (G) Superimposition of the ACE2-bound SARS-CoV-2 RBD structure (PDB 7DMU) on the SARS-CoV-2 B.1.427/B.1.429 S cryoEM structure show that L452R points away from the interface with ACE2.
CryoEM structure of the SARS-CoV-2 B.1.427/B.1.429 S ectodomain trimer. (A) Structure of the S trimer (surface rendering) bound to the S2M11 and S2L20 Fabs (ribbons) in two orthogonal orientations. SARS-CoV-2 S protomers are colored pink, cyan, and gold, whereas the S2L20 Fab heavy and light chains are colored dark and light green, respectively, and the S2M11 Fab heavy and light chains are colored dark and light gray, respectively. Only the Fab variable domains are resolved in the map. N-linked glycans are rendered as dark blue spheres. (B) Zoomed in view of the S2M11-bound RBD with R452 shown in ball and stick representation. (C) Zoomed in view of the S2L20-bound NTD with disordered N terminus, supersite β-hairpin and loop regions shown as dashed lines. (D) Superimposition of the CT-P59–bound SARS-CoV-2 RBD structure (PDB 7CM4) on the SARS-CoV-2 B.1.427/B.1.429 S cryoEM structure show that R452 would sterically clash with the mAb. (E) Superimposition of the LY-CoV555–bound SARS-CoV-2 RBD structure (PDB 7KMG) on the SARS-CoV-2 B.1.427/B.1.429 S cryoEM structure show that L452R would sterically clash with the mAb. (F) Superimposition of the S2X333-bound SARS-CoV-2 S structure (PDB 7LXW) on the SARS-CoV-2 B.1.427/B.1.429 S cryoEM structure reveals that most of the NTD antigenic supersite epitope residues are disordered. (G) Superimposition of the ACE2-bound SARS-CoV-2 RBD structure (PDB 7DMU) on the SARS-CoV-2 B.1.427/B.1.429 S cryoEM structure show that L452R points away from the interface with ACE2.
The S13I/W152C mutant NTD escaped all NTD-directed neutralizing antibodies, even though the non-neutralizing anti-NTD antibody retained full binding. The researchers demonstrated that the C15/C136 disulfide bond holds the N-terminus of the NTD to the rest of the same domain.
With the S131 mutation in the NTD, cleavage at S13-Q14 allows the C136 residue to be bound by cysteine from the expression media. A similar cysteinylation occurs with W152C. This disrupts the C15-C136 disulfide bond, thus introducing disorder into the N terminal end of the B.1.427/B.1.429 S protein.
Though these individual mutations lead to cysteinylation at C136 and W152, respectively, this is not the case when both occur together. In this situation, a disulfide bond forms between C136 and W152C in the corresponding B.1.427/B.1.429 variant. This introduces profound disorder in the β-hairpin of the antigenic supersite, which houses the W152C position, as this new disulfide bond with C136 causes residues to shift by over 20 Å.
Binding to the NTD by neutralizing antibodies was reduced more with the former than the latter mutation. These findings support the hypothesis that clinically important immune-evading mutations of the S protein emerge in vivo, thereby indicating that the virus uses an indirect strategy to escape neutralizing antibodies specific to the NTD.
What are the implications?
The investigators suggest that the L452R mutation was acquired by multiple lineages as a result of positive selection following the exposure of the virus to RBD-targeting neutralizing antibodies. The effect is immune evasion by altered binding of several RBD-specific antibodies.
However, many variants of the virus in current circulation affect the NTD, including the antigenic supersite, which indicates that this region is under strong selection pressure by host immune responses. In fact, chronic SARS-CoV-2 infections in individuals with weakened immunity have produced variants with deletions within the NTD antigenic supersite. In addition, in vitro observations have found that escape mutations in this region undergo selection that reduces the binding of the virus by convalescent sera or monoclonal antibodies.
“These findings demonstrate that the S13I and W152C mutations found in the B.1.427/B.1.429 S variant are jointly responsible for escape from NTD-specific mAbs, due to deletion of the SARS-CoV-2 S two N-terminal residues and overall rearrangement of the NTD antigenic supersite.”
In the near future, the B.1.427/B.1.429 lineages may acquire the E484K mutation, which confers broad escape capabilities against multiple RBD-directed monoclonal antibodies. This same immune evasion characteristic has appeared in the B.1.351, P.1, B.1.526, and even the B.1.1.7 variants.
“Understanding the newfound mechanism of immune evasion of the emerging variants, such as the signal peptide modification described herein, is as important as sequence surveillance itself to successfully counter the ongoing pandemic.”