Scientists from the UK and US have recently investigated immunological consequences of secondary Streptococcus pneumoniae (S. pneumoniae) infection in coronavirus disease 2019 (COVID-19) patients. The study reveals that S. pneumoniae, which is a Gram-positive bacterium responsible for pneumonia, may modulate the host immune system to facilitate the immune escape of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The study is currently available on the medRxiv* preprint server.
Bacterial colonization in the upper respiratory tract is considered to be one of the major risk factors of COVID-19. In this context, studies have shown that the co-occurrence of invasive pneumococcal disease in COVID-19 patients increases the risk of mortality.
Mechanistically, an interaction between S. pneumoniae and respiratory viruses, such as influenza virus and respiratory syncytial virus, in the upper respiratory tract increases the risk of secondary pneumococcal pneumonia. However, in COVID-19 patients, the incidence of pneumococcal pneumonia has been observed only rarely. This could be due to low bacterial transmission owing to non-pharmacological control measures or low bacterial detection because of excessive use of empiric antibiotics.
From the immunological perspective, studies have shown that during viral infection, bacterial colocalization in the upper respiratory tract can lead to modulation of host immune responses, which in turn can facilitate immune evasion of the virus.
In the current study, the scientists have investigated the impact of S. pneumoniae and SARS-CoV-2 co-infection on host adaptive immune responses.
Study design
The study was conducted on healthcare workers with asymptomatic or mildly symptomatic COVID-19 and hospitalized COVID-19 patients with moderate-to-severe disease. In both groups, the scientists estimated the prevalence of S. pneumoniae and SARS-CoV-2 co-infection and its impact on COVID-19 severity, immune responses, and inflammation.
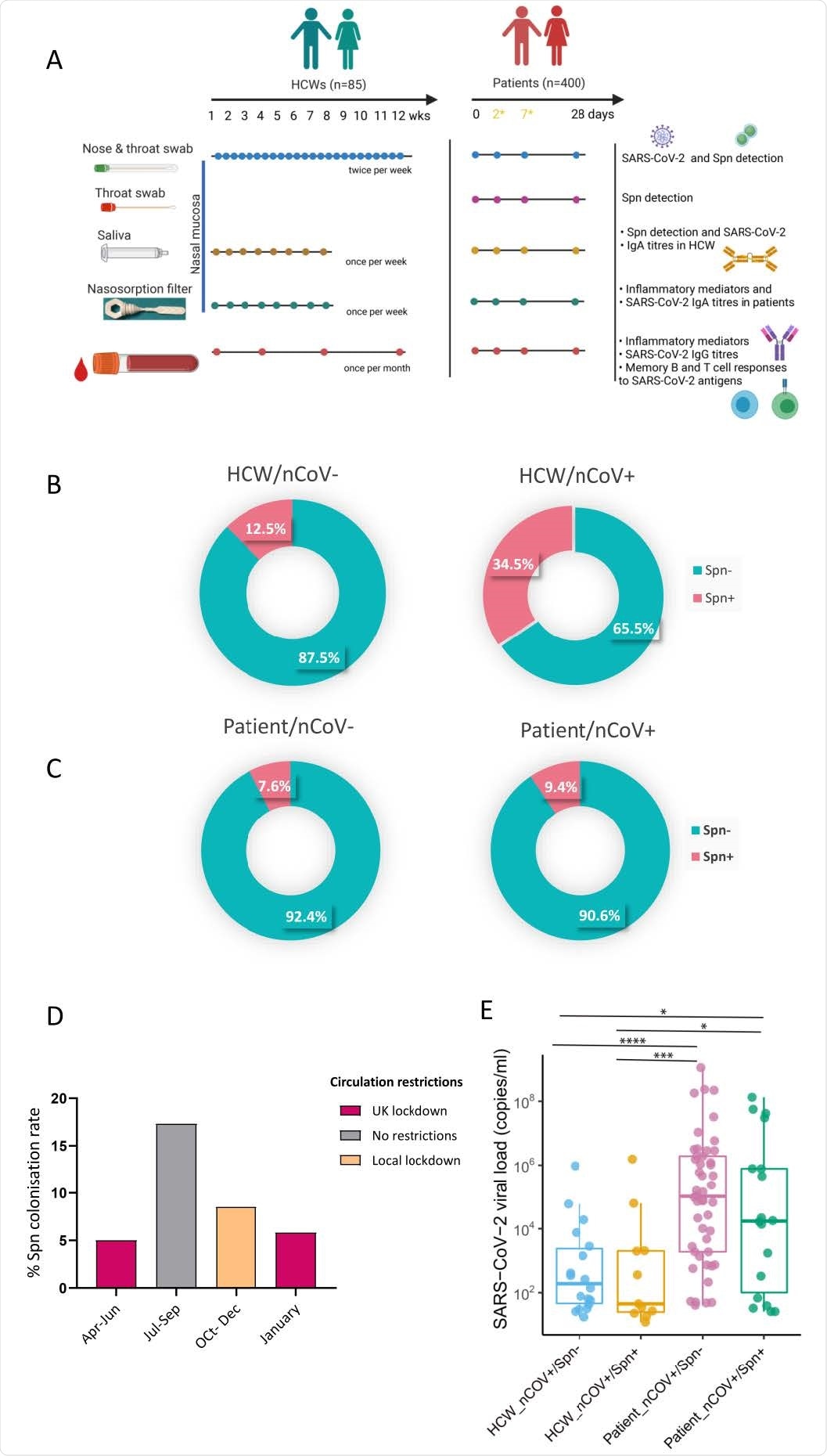
Prevalence of pneumococcal colonization amongst SARS-CoV-2 positive and negative HCWs and patients. A) Experimental design of the study with sample type, sample collection schedule and measurable per sample type depicted for both HCW and patient cohorts. In patient cohort, day2 and day7 samples were collected only if individuals who were hospitalized. B-C) Doughnut charts showing the percentage of pneumococcal prevalence in B) HCWs (n=85) and C) patients (n=400), infected and non-infected with SARS-CoV-2. Fisher’s exact test was used to compare percentages. D) Percentage of pneumococcal colonization rate detected in patient cohort during calendar periods of different circulation restrictions rules applied. 5% (6/119) from April- June, 17.3% (13/75) from July to September, 8.5% (13/154) from October to December and 5.8% (3/52) in January. E) Levels of viral load, expressed as RNA copies/ml as detected by Genesig RT-qPCR in HCW non-colonised (n=19, light blue) and Spn-colonised (n=10, yellow) and patient non-colonised (n=73, lilac) and Spn-colonised (n=19, green).
Importance observations
Of 85 enrolled healthcare workers, 29 tested positive for SARS-CoV-2 and 17 tested positive for S. pneumoniae. The prevalence of S. pneumoniae infection was significantly higher in SARS-CoV-2 positive participants than SARS-CoV-2 negative participants.
Similarly, of 400 enrolled hospitalized patients, 255 tested positive for SARS-CoV-2 and 35 tested positive for S. pneumoniae. However, no significant difference in S. pneumoniae prevalence was observed between SARS-CoV-2 positive and negative patients. Importantly, no effect of S. pneumoniae co-infection on upper airway viral load and disease severity was observed among participants.
Mucosal antibody responses to SARS-CoV-2
Since SARS-CoV-2 primarily attacks upper airway epithelial cells, IgA antibody-induced mucosal antiviral immunity is expected to play a vital role in the early elimination of the virus.
To characterize mucosal immune responses, the scientists measured the levels of IgA-specific anti-SARS-CoV-2 antibodies in saliva and nasal samples. The findings revealed that in SARS-CoV-2 positive healthcare workers, S. pneumoniae co-infection led to a significant reduction in mucosal IgA responses to SARS-CoV2. However, in SARS-CoV-2 positive hospitalized patients, this change was not very prominent. Regarding systemic antibody responses, SARS-CoV-2 positive healthcare workers with S. pneumoniae co-infection showed a trend toward lower induction of IgG-specific immune responses to SARS-CoV-2. However, in hospitalized patients with S. pneumoniae co-infection, significantly lower levels of IgG-specific anti-SARS-CoV-2 antibodies were detected in serum samples.
For mechanistic evaluation, the scientists conducted a series of co-colonization experiments with live-attenuated influenza virus and S. pneumoniae.
The findings revealed that S. pneumoniae infection occurred prior to influenza virus exposure led to suppression of IgA immune responses to influenza virus. In contrast, no such alteration was observed when viral exposure occurred before S. pneumoniae infection.
Memory B cell and T cell responses to SARS-CoV-2
To evaluate long-term humoral immunity, the scientists isolated SARS-CoV-2 specific memory B cells from peripheral blood mononuclear cells of participants. The findings revealed that co-infection with S. pneumoniae and SARS-CoV-2 resulted in reduced memory B cell response to SARS-CoV-2 antigens in both healthcare workers and patients.
In hospitalized patients co-infected with S. pneumoniae, significantly lower levels of CD4+ T cell response to SARS-CoV-2 spike, nucleocapsid, and S1 antigens were observed. A similar trend was observed for CD8+ T cell responses to different SARS-CoV-2 antigens.
Inflammatory response
The scientists measured the levels of 30 cytokines in nasal and serum samples during acute infection to investigate SARS-CoV-2 induced inflammatory responses. The findings revealed that in hospitalized patients, S. pneumoniae co-infection resulted in a reduced serum inflammatory profile. However, in both colonized or non-colonized healthcare workers, the level of inflammation was lower than in hospitalized patients.
In nasal samples, interestingly, healthcare workers showed reduced levels of certain cytokines that are associated with T cell maturation and differentiation. Importantly, in both healthcare workers and hospitalized patients, S. pneumoniae infection did not cause a variation in cytokines associated with T cell proliferation and activation.
Study significance
The study highlights the significance of S. pneumoniae infection in COVID-19 patients in modulating host adaptive immunity against SARS-CoV-2, which in turn can facilitate viral immune evasion. As mentioned by the scientists, colocalization of S. pneumoniae with SARS-CoV-2 may increase the risk of long-COVID or SARS-CoV-2 reinfection.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources