Researchers have just established a structural basis for two spike glycoprotein epitopes of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that are restricted to human leukocyte antigen (HLA) A2 molecules. More specifically, they are involved in the presentation of antigenic peptides to specific T lymphocytes and recognized by public and private T cell receptors. The research is currently available on the bioRxiv* preprint server.
Study: Structural basis for recognition of two HLA-A2-restricted SARS-CoV-2 spike epitopes by public and private T cell receptors. Image Credit: Meletios Verras / Shutterstock
Although current vaccines against coronavirus disease (COVID-19) caused by SARS-CoV-2 are effective against that virus (notwithstanding a lower neutralization ability against specific viral variants), a holy grail would be a pan-coronavirus vaccine that could protect against infection with potential emergent coronaviruses (and not only SARS-CoV-2).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
To achieve that, more knowledge in fundamental immunology is needed. For example, we have an abundance of structural data on neutralizing antibodies, but not so much regarding T cells that have a pivotal role in combatting the infection and forming a long-term immune memory response.
In other words, while we have a rather detailed picture of the B cell response and antibody production when the human organism is confronted with SARS-CoV-2, we lack specific information on specific T cell receptors (TCRs) for SARS-CoV-2 (but also other coronaviruses) that are bound to their peptide–MHC targets and that basically orchestrate the antiviral response.
In a majority of T cell responses, TCR repertoires that are evoked by a particular antigenic epitope differ between individuals (also known as private T cell responses); conversely, certain other epitope-specific TCR repertoires contain TCRs that are habitually encountered in multiple unrelated individuals (also known as public T cell responses).
In this new study, a research group from the University of Maryland in the US, University of South China and National Research Center for Hematology in Moscow (Russia) aimed to take a deep look into public and private TCRs in order to inform prospective vaccine design efforts.
Structure, binding and computational analysis
In short, by using a state-of-the-art methodology and computational analysis, this study has revealed crystal structures of one public and one private SARS-CoV-2 TCR. More specifically, these were immunodominant yet variable epitope (YLQ) and a conserved but less commonly targeted epitope (RLQ).
These specific TCRs were then tested for cross-reactivity with other human coronaviruses (most notably original SARS-CoV and MERS, but also seasonal coronaviruses such as HKU1, NL63 and OC43) with the use of peptides homologous to the YLQ and RLQ epitopes of SARS-CoV-2.
The ability of the aforementioned TCRs YLQ and RLQ to recognize two natural epitope variants found in the GISAID database and representing the most common mutations among SARS-CoV-2 spike glycoprotein sequences has also been tested. All interactions were tested by utilizing detailed structural analyses.
High specificity of human T-cell receptors
In combination with specific binding experiments and meticulous computational analysis, structural determination has underscored a quite exquisite specificity of human TCRs towards two specific T cell epitopes from SARS-CoV-2, which is the crux of this study.
Furthermore, the basis for selection of particular human T Cell receptor alpha and beta variable germline genes by the public, but not the private TCR was also revealed in the structures, as well as the propensity of both TCRs to discern natural variants of YLQ and RLQ – but not homologous epitopes from human seasonal coronaviruses, which was an exciting finding.
Finally, the researchers have also found that two natural variants of the YLQ and RLQ epitopes harboring the most frequent epitope mutations in the GISAID database activated T cells in functional assays almost as efficiently as the wild-type epitopes.
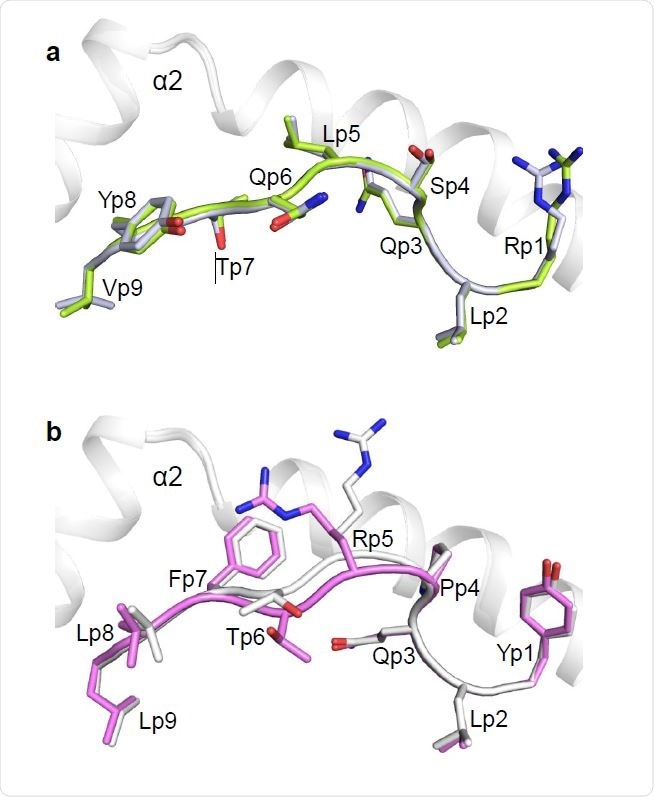
Conformations of RLQ and YLQ peptides bound to HLA-A2. (a) Side view of two superposed RLQ–HLA-A2 molecules in the asymmetric unit of the crystal. Carbon atoms of the superposed RLQ peptides are green or gray; nitrogen atoms are blue; oxygen atoms are red. HLAA2 is gray. Residue labels for RLQ are aligned with the α-carbon atom of the respective residue. (b) Side view of two superposed YLQ–HLA-A2 molecules in the asymmetric unit of the crystal. Carbon atoms of the superposed YLQ peptides are violet or gray.
Towards a universal vaccine
By laying out the mechanistic basis of TCR targeting of an immunodominant yet variable site, as well as a conserved and less commonly targeted site, this important study gave us useful information for our further efforts to rational vaccine design and optimization efforts.
“An additional consideration in pancoronavirus vaccine design is the effective induction of T cell responses to epitopes that are conserved across coronaviruses, such as the RLQ epitope”, say study authors in this bioRxiv paper.
“This point is underscored by this study, where through structural determination, binding experiments, and computational analysis, we have highlighted the exquisite specificity of human TCRs that target two T cell epitopes from SARS-CoV-2”, they add.
Hence, these findings are another step forward towards the development of long-lasting vaccines that will also show a cross-protective immunity against many different (and potentially emerging) coronaviruses.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources