The coronavirus disease 2019 (COVID-19), which is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has devastated the world’s healthcare and economic systems. As of August 6, 2021, SARS-CoV-2 has caused over 4.3 million deaths and infected over 200 million people worldwide. While most patients recover, persistent symptoms have been repeatedly described.
 Study: Long-term perturbation of the peripheral immune system months after SARS-CoV-2 infection. Image Credit: levin_design / Shutterstock.com
Study: Long-term perturbation of the peripheral immune system months after SARS-CoV-2 infection. Image Credit: levin_design / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
In most cases, COVID-19 presents as asymptomatic or with mild symptoms. Aside from these cases, many people infected with COVID-19 have developed moderately severe symptoms like fever, cough, headache, tiredness, diarrhea, and disturbances of smell and taste.
A significant minority of individuals with COVID-19 will proceed to develop severe or critical disease. This is particularly true for individuals with underlying comorbidities such as obesity, diabetes, or cardiovascular disease, or who are older in age.
Among those who recover from COVID-19, which comprises the vast majority of cases, some report persistent symptoms for months, independent of the severity of their symptoms. This condition is often referred to as ‘long COVID,’ and has been reported to affect up to two-thirds of discharged patients, in some studies. While most of the COVID-19 long-haulers describe continuing tiredness, muscle weakness, sleep disruption, anxiety, and depression, those who survived severe acute lung injury often have residual impairment of respiratory function and abnormal findings on subsequent chest X-rays.
The commonality of post-infectious syndromes after COVID-19, Ebola virus, and COVID-19 indicates that these are underlaid by a persistent abnormality in immune regulation in these individuals.
In earlier studies, flow cytometry of peripheral blood samples from COVID-19 patients showed that both innate and adaptive immune cells change in their frequency. For instance, recovered patients had altered CD4+ and CD8+ T cell activation and exhaustion markers. Classical CD14+ monocytes, which belong to the innate immune system, were increased and showed inflammatory profiles. In those with a history of severe COVID-19, plasmacytoid Dendritic Cells (pDCs) were decreased, while effector CD8+ T cells were higher than in healthy controls.
Current study
The current study was carried out in Australia, which was under strict containment measures in force at the time at which the study was conducted. These measures largely prevented the re-infection of recovered COVID-19 patients.
Three aspects of the immune responses to COVID-19 were evaluated in this study. These include the production of antibodies to the viral spike (S) and receptor-binding domain (RBD), multiple immunological phenotypic parameters, as well as transcriptomics. The tested cohorts included individuals who had recovered from both mild/moderate or severe/critical COVID-19, as well as healthy controls.
A total of 69 cases were tested at 12, 16, and 24 weeks from the first positive SARS-CoV-2 test. Of the cases, 50 had a history of mild COVID-19, while 6, 7, and 6 had moderate, severe, or critical disease, respectively. These results were compared with 14 seronegative healthy controls.
What were the antibody findings?
The study shows that antibodies were robustly induced by COVID-19 in samples collected six months after recovering from the infection. More specifically, the immunoglobulin G (IgG) antibody titers to the S and RBD antigens covered a wide range, but remained stable over time, with a tendency to go down as time increased.
However, IgM and IgA titers decreased over time. Anti-RBD IgG3 and IgM titers showed a more rapid decline over time as compared to antibodies against the S protein.
The anti-S protein IgG titers were higher in those who had recovered from severe or critical disease as compared to milder cases, either at all time points or at 24 weeks only. Similar differences were observed with anti-RBD antibodies, but only at 24 weeks. The titers of total anti-S protein and anti-RBD IgG antibodies remained correlated throughout.
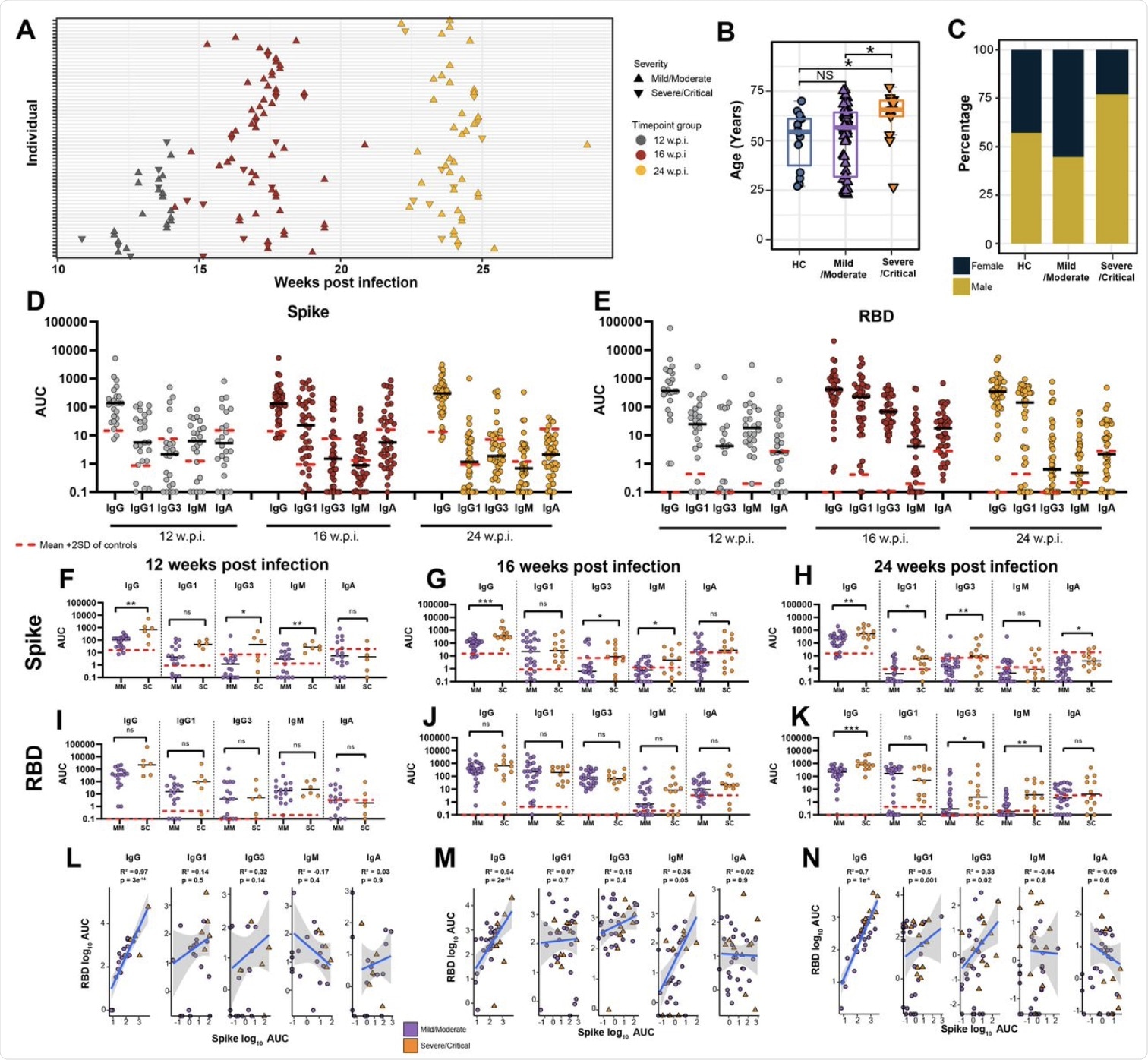 (A) Blood sample collection timepoints. (B) Age and (C) sex distribution of healthy controls (HC) in comparison to mild/moderate and severe/critical COVID-19 convalescents. (D) Anti-Spike and (E) anti-RBD specific IgG, IgG1, IgG3, IgM and IgA titres at 12, 16, and 24 w.p.i. End point titers are reported as area under the curve (AUC). The mean is denoted by the horizontal black lines. Seronegative samples were assigned a value of 0.1. Red dashed lines represent the mean AUC + 2 SD in HC for each isotype. (F-K) Antibody titres subdivided by disease severity. (L-M) Pearson correlations between anti-Spike and anti-RBD antibody subclass titres at each timepoint. Statistical significance was assessed in (B,F-K) using Wilcoxon Rank Sum Tests. ns= non-significant. * P < 0.05, ** P < 0.01, *** P < 0.001.
(A) Blood sample collection timepoints. (B) Age and (C) sex distribution of healthy controls (HC) in comparison to mild/moderate and severe/critical COVID-19 convalescents. (D) Anti-Spike and (E) anti-RBD specific IgG, IgG1, IgG3, IgM and IgA titres at 12, 16, and 24 w.p.i. End point titers are reported as area under the curve (AUC). The mean is denoted by the horizontal black lines. Seronegative samples were assigned a value of 0.1. Red dashed lines represent the mean AUC + 2 SD in HC for each isotype. (F-K) Antibody titres subdivided by disease severity. (L-M) Pearson correlations between anti-Spike and anti-RBD antibody subclass titres at each timepoint. Statistical significance was assessed in (B,F-K) using Wilcoxon Rank Sum Tests. ns= non-significant. * P < 0.05, ** P < 0.01, *** P < 0.001.
Immune cell finding
The researchers explored ten types of immune cells that were separated in each category on functional markers. These immune cells showed marked alterations, particularly at 12 and 16 weeks after the infection. While the differences in these cellular phenotypes were most prominent at 12 weeks, only some persisted to 24 weeks.
For instance, lymphocyte counts remained depressed for up to 16 weeks following COVID-19 infection, even as CD3+ T cells and CD19+ B cells increased. Additionally, CD38+CD27+ memory B cells showed a significant rise.
Conversely, both the CD4+ and CD8+ T cells were reduced at 12 and 16 weeks. This was accompanied by a reduction in CD4+ effector memory (EM) pools, as well as migratory central memory (CM) CD4+ T cells throughout the study period.
Natural killer (NK) cells were also increased following the COVID-19 diagnosis, both total and tissue migratory phenotypes. Total granulocytes, including low density (LD) neutrophils, were increased for up to 16 weeks. Among the latter, those expressing CXCR3+, which indicates their active migration into injured tissues, were higher at 12 weeks and returned to normal by 16 weeks. Notably, CD14+CD16+ neutrophils remained low at both time points.
At 12 weeks, monocytes levels remained consistent. However, tissue-homing monocytes bearing the CXCR3+ marker, both activated and regulatory monocytes, were elevated.
Antibody titers were correlated with granulocytes, NK CD16+ cells, and CD4+ T cells, thereby suggesting that they both mirror disease severity and that each of these cells may help boost the T cell response.
T helper cells type 9 (Th9) declined throughout the study period; however, Th2/22 cells rose at 16 weeks. Since all of these subsets are epithelial-homing cells, these alterations reflect a dysregulated lung mucosal repair process. This observation could also indicate that these cells left the peripheral circulation on a large scale in order to enter damaged sites.
Th17 and Th22 cells both showed higher proportions of mature T cells. The frequencies of these cells were consistently reduced at all time points, while their relative proportions remained constant. These may therefore be implicated in antiviral immune responses.
Memory Th12/Th22 cells were also increased at 12 weeks, thus indicating that these cells were establishing immune memory focused on tissue healing. Some subsets of T follicular helper (Tfh) cells were elevated, such as Tfh1 and Tfh2/22.
T regulatory cells (Tregs) that modulate these T cell lineages were increased in their naïve form throughout the course of the study; however, mature cells were consistently reduced up to 16 weeks. The changes in various Treg subsets indicate that the follicular regulatory T cell lineages (TfhR), which regulate both Tfh and B cell help, were reduced during the same period; however, the levels of these cells returned to normal by 24 weeks. The signal recruiting Th cells into inflamed or damaged tissue may therefore continue to operate for months after the initial infection.
Taken together, this immune subset apparently responded to the early infection. As the virus migrated into the tissue, the TfhR cells increased, thereby indicating their role in B cell help within the lymphoid germinal centers. These two phases are required for a robust antiviral B cell response.
Both ThR2/22 and TfhR2/22 subsets were positively associated with anti-SARS-CoV-2 S protein and RBD IgG levels. This suggests an antibody-regulatory function for this epithelial homing lineage.
Transcriptomics
RNA sequencing showed significant changes in the way genes were expressed following infection for up to 24 weeks following infection, irrespective of disease severity. Genes involving ribosome biosynthesis, oxidative phosphorylation, as well as platelet activation and signaling, were downregulated for up to 16 weeks. At 24 weeks, only the complement activation pathway was downregulated.
Several inflammatory pathways were upregulated in COVID-19 convalescents. Further analysis showed that these long-term disruptions were not explained by changes in any given immune cell population.
Another approach using blood transcriptional modules (BTMs) showed that while more recovering patients showed increasing BTM activity shifting towards the healthy control baseline profile, a set of patients continued to show persistent dysregulation, even at six months.
“These data suggest ongoing inflammatory responses and immune dysregulation in COVID-19 convalescents weeks-to-months after infection.”
What are the implications?
In summary, our integrated network analysis reveals a complex interplay of relationships between circulating immune cell populations, transcriptional dysregulation, and humoral immune responses in COVID-19 convalescent patients and provides a resource for further exploration and investigation of these relationships.”
Overall, many convalescents remained antibody-positive, reacting to both the S and RBD antigens for up to six months after infection. Antibody titers were higher in those recovering from severe disease, which mirrors recent studies investigating the neutralizing activity of serum in these patients.
Interestingly, IgM and IgG1 target the S1 domain of the S protein, thus impacting virus binding to the host cell. This is the focus of activity for most neutralizing antibodies and monoclonal antibodies.
Conversely, IgG3 antibodies bind to the S2 domain with higher affinity, which may prevent virus entry and syncytia formation. Thus, further research may focus on the relative roles of these antibodies in virus neutralization, as well as inhibiting virus infection by preventing the membrane fusion triggered by viral binding.
Similarly, some immune cell subsets continued to show marked alterations relative to the baseline, even at six months. Lymphopenia persisted until 16 weeks, which was accompanied by changes in other T cell subsets. Memory B cells also appeared to remain activated and exhausted at three months.
Naïve Tregs appeared to expand, perhaps to restore their numbers that were previously depleted by inflammation and tissue damage. This supports current views that Tregs are required both to modulate the immune response and enhance tissue repair processes.
Increased TEMRA Tregs are considered to be a sign of T cell exhaustion, but could instead represent cytotoxic, migratory, and tissue repair activity in a subset of polyfunctional effector Tregs. In fact, the researchers comment, their presence may “suggest a competition between classical immune suppression and tissue repair by these cells in response to tissue damage in COVID-19 convalescents.”
The differences in how and when such cellular and transcriptional changes revert to normal may explain long COVID development; however, this requires further study. Moreover, persistent dysregulation is perhaps an accompaniment of impaired humoral responses, and therefore of shorter protective immunity.
“These changes to the peripheral immune system could have implications for how individuals recovering from infection respond to other challenges encountered in this period and persistent immune activation may also exacerbate other chronic conditions.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Ryan, F. J., Hope, C. M., Masavuli, M. G., et al. (2021). Long-term perturbation of the peripheral immune system months after SARS-CoV-2 infection. medRxiv. doi:10.1101/2021.07.30.21261234. https://www.medrxiv.org/content/10.1101/2021.07.30.21261234v1.
- Peer reviewed and published scientific report.
Ryan, Feargal J., Christopher M. Hope, Makutiro G. Masavuli, Miriam A. Lynn, Zelalem A. Mekonnen, Arthur Eng Lip Yeow, Pablo Garcia-Valtanen, et al. 2022. “Long-Term Perturbation of the Peripheral Immune System Months after SARS-CoV-2 Infection.” BMC Medicine 20 (1). https://doi.org/10.1186/s12916-021-02228-6. https://bmcmedicine.biomedcentral.com/articles/10.1186/s12916-021-02228-6.