Researchers in the United States, Australia and the UK have developed a novel method that enables the evolutionary ancestry (phylogeny) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) to be comprehensively searched for recombinant lineages.
The SARS-CoV-2 virus is the agent responsible for the ongoing COVID-19 pandemic that continues to pose a threat to global public health and the worldwide economy.
Recombination is a key contributor to the genetic variation that occurs in SARS-CoV-2. By combining genetic material from diverse genomes, recombination can generate novel sets of mutations that have potentially important phenotypic effects.
The COVID-19 pandemic has driven an unprecedented surge in pathogen genome sequencing and data sharing. Unfortunately, this has highlighted the limitations of current software systems, with large genomic datasets often exceeding the capacity of existing platforms and crippling the real-time analysis of viral recombination.
Now, researchers from the University of California in Santa Cruz, the Wellcome Genome Campus in Cambridge, UK, and the Australian National University in Canberra have developed a platform called Recombination Inference using Phylogenetic PLacEmentS (RIPPLES) that can search the entire phylogenetic tree of SARS-CoV-2 and detect recombination both within and between different lineages.
The team identified 606 recombination events and estimated that approximately 2.7% of the SARS-CoV-2 genomes sequenced have recombinant ancestry.
Yatish Turkahia and colleagues say that as SARS-CoV-2 populations accumulate genetic diversity and co-infect hosts that harbor other species of viruses, recombination will play an increasingly large role in generating functional genetic diversity.
“RIPPLES is therefore poised to play a primary role in detecting novel recombinant lineages and quantifying their impacts on viral genomic diversity as the pandemic progresses,” they write.
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Monitoring of recombination is essential
The accurate and timely characterization of recombination is critical to understanding the phylogeny of SARS-CoV-2 in human, agricultural, and wild animal populations.
However, the vast amount of genomic data generated during the COVID-19 pandemic has overwhelmed existing analysis platforms and hindered the real-time analysis of viral recombination.
New approaches for detecting and characterizing recombinant haplotypes of SARS-CoV-2 are needed to assess new variant genome sequences as quickly as they become available, says Turakhia and colleagues.
“Such rapid turnaround is essential for driving an informed and coordinated public health response to novel SARS-CoV-2 variants,” they write.
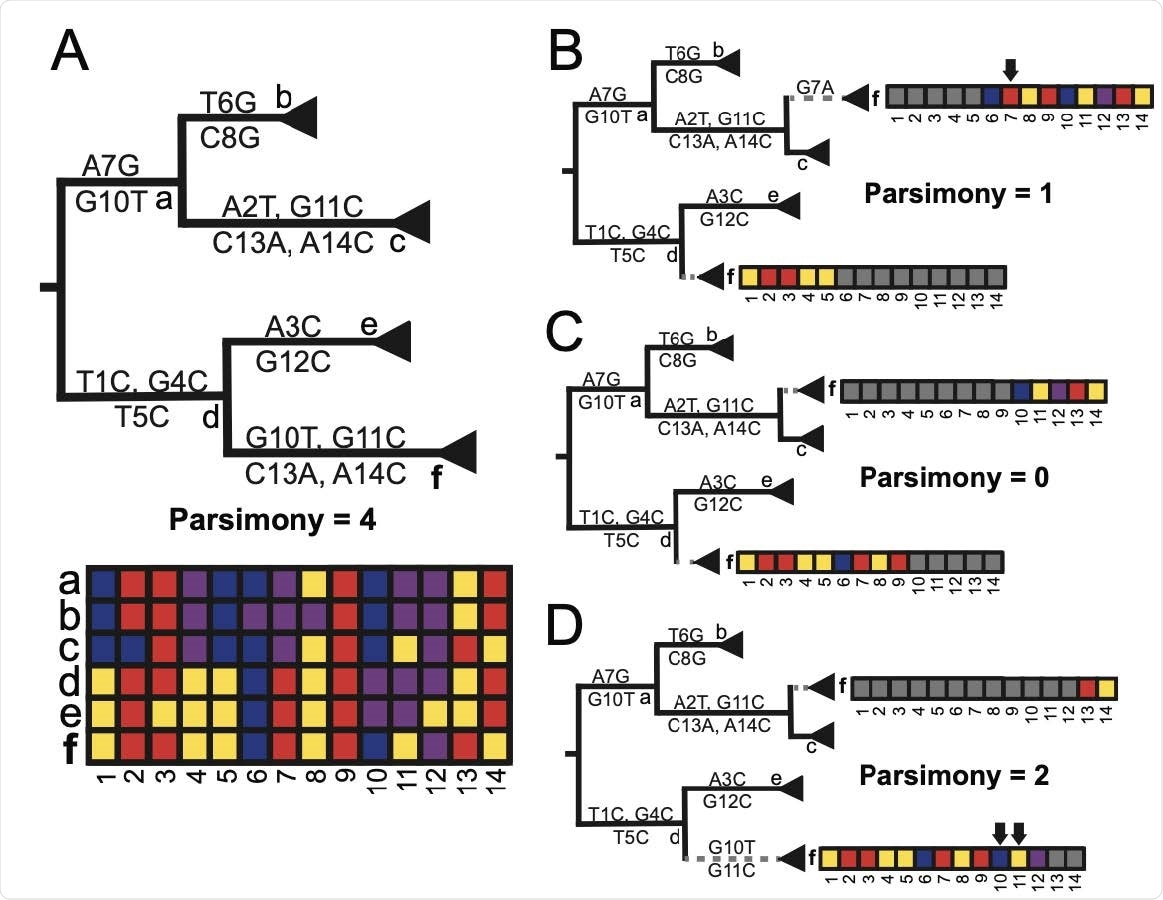
RIPPLES exhaustively searches for optimal parsimony improvements using partial interval placements. (A): A phylogeny with 6 internal nodes (labeled a-f), in which node f is the one being currently investigated as a putative recombinant. The initial parsimony score of node f is 4, according to the multiple sequence alignment below the phylogeny, which displays the variation among samples and internal nodes. Note that internal nodes may not have corresponding sequences in reality, but test for recombination using reconstructed ancestral genomes. (B-D): Three partial placements given breakpoints are shown with their resulting parsimony scores. Arrows mark sites that increase the sum parsimony of the two partial placements of f. The optimal partial placement and breakpoint prediction for node f is in the center (C), with one breakpoint after site 9 and with partial placements both as a sibling of node c and as a descendant of node d.
What did the researchers do?
Now, the researchers have developed the novel RIPPLES method, which can detect recombination in pandemic-scale phylogenies.
“Our extensively optimized implementation of RIPPLES allows it to search the entire phylogenetic tree and detect recombination both within and between SARS-CoV-2 lineages without a priori defining a set of lineages or clade-defining mutations,” say the researchers.
“This is a key advantage of our approach relative to other methods that cope with the scale of SARS-CoV-2 datasets by reducing the search space for possible recombination events.”
Ripples detected simulated recombinants with 93% sensitivity in just 6.25 minutes across a global phylogeny containing more than 1.6 million SARS-CoV-2 sequences.
The team identified 606 unique recombination events, for which the combined total of descendant samples was 43,163.
This means that around 2.7% of the genome’s samples were inferred to belong to detectable recombinant lineages.
The researchers say that since the recombination events that occur between genetically similar viral lineages are challenging to detect, this figure is expected to be a potentially significant underestimate of the overall frequency of recombination.
“The RIPPLES estimate is likely conservative with respect to the global frequency of recombination in the SARS-CoV-2 population,” says the team.
“The spike protein is a primary location of functional novelty for viral lineages”
RIPPLES revealed that recombination breakpoints occurred disproportionately in the SARS-CoV-2 spike protein – the main structure the virus uses to bind to and infect cells.
“The spike protein is a primary location of functional novelty for viral lineages as they adapt to transmission within and among human hosts,” writes Turakhia and colleagues.
The researchers say that the discovery of the excess of recombination events around spike, as well as the relatively high levels of recombinants that are currently circulating, highlights the importance of monitoring the evolution of new viral lineages using real-time analyses.
“To facilitate real-time analysis of recombination among tens of thousands of new SARS-CoV-2 sequences being generated by diverse research groups worldwide each day, RIPPLES provides an option to evaluate evidence for recombination ancestry in any user-supplied samples within minutes,” they write.
“RIPPLES, therefore, opens the door for rapid analysis of recombination in heavily sampled and rapidly evolving pathogen populations, as well as providing a tool for real-time investigation of recombinants during a pandemic,” concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Turkahia Y, et al. Pandemic-Scale Phylogenomics Reveals Elevated Recombination Rates in the SARS-CoV-2 Spike Region. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.08.04.455157, https://www.biorxiv.org/content/10.1101/2021.08.04.455157v1
- Peer reviewed and published scientific report.
Turakhia, Yatish, Bryan Thornlow, Angie Hinrichs, Jakob McBroome, Nicolas Ayala, Cheng Ye, Kyle Smith, et al. 2022. “Pandemic-Scale Phylogenomics Reveals the SARS-CoV-2 Recombination Landscape.” Nature 609 (7929): 994–97. https://doi.org/10.1038/s41586-022-05189-9. https://www.nature.com/articles/s41586-022-05189-9.