A team of US-based scientists has recently conducted a robust high-throughput screening to identify potential entry inhibitors of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The screening identifies calpeptin as a potent and highly specific inhibitor of SARS-CoV-2 and its variants. The study is currently available on the bioRxiv* preprint server.
SARS-CoV-2, the causative pathogen of coronavirus disease 2019 (COVID-19), has infected more than 202 million individuals and claimed 4.29 million lives globally. Despite non-pharmacological control measures and vaccines, the pandemic trajectory is still rising.
Several antiviral medicines have been repurposed as therapeutic interventions, and monoclonal antibodies targeting the viral spike protein have been developed.
Although the effectiveness of repurposed medicines is still uncertain, combination therapies with monoclonal antibodies are showing promising results, especially in hospitalized COVID-19 patients with severe disease.
It is now well-established that SARS-CoV-2 entry into host cells is initiated by the interaction of viral spike with host cell angiotensin-converting enzyme 2 (ACE2), followed by fusion of viral envelope – host cell membrane. To prevent SARS-CoV-2 infection at early stages, it is, thus, essential to develop or identify potential small molecules that can interfere with the spike – ACE2 interaction.
In the current study, the scientists have screened several unique libraries to identify small molecules that can be repurposed to inhibit the SARS-CoV-2 entry process.
Specifically, they have screened the ReFRAME library of 13,136 small molecules that have already undergone preclinical testing or are currently under clinical development. Additionally, they have screened 15,000 small molecules from two other libraries and 400 molecules with known or suspected antiviral activity.
For the validation of identified molecules, they have conducted a series of mechanistic assays, including whole-cell SARS-CoV-2 infectivity assay.
Identification of potent candidate molecule
The screening of around 15,000 clinically relevant small molecules led to the identification of calpeptin as the most potent candidate molecule targeting the SARS-CoV-2 entry process. Calpeptin is a cell-permeable inhibitor of a group of proteases, including calpain I, calpain II, cathepsin L, and cathepsin K.
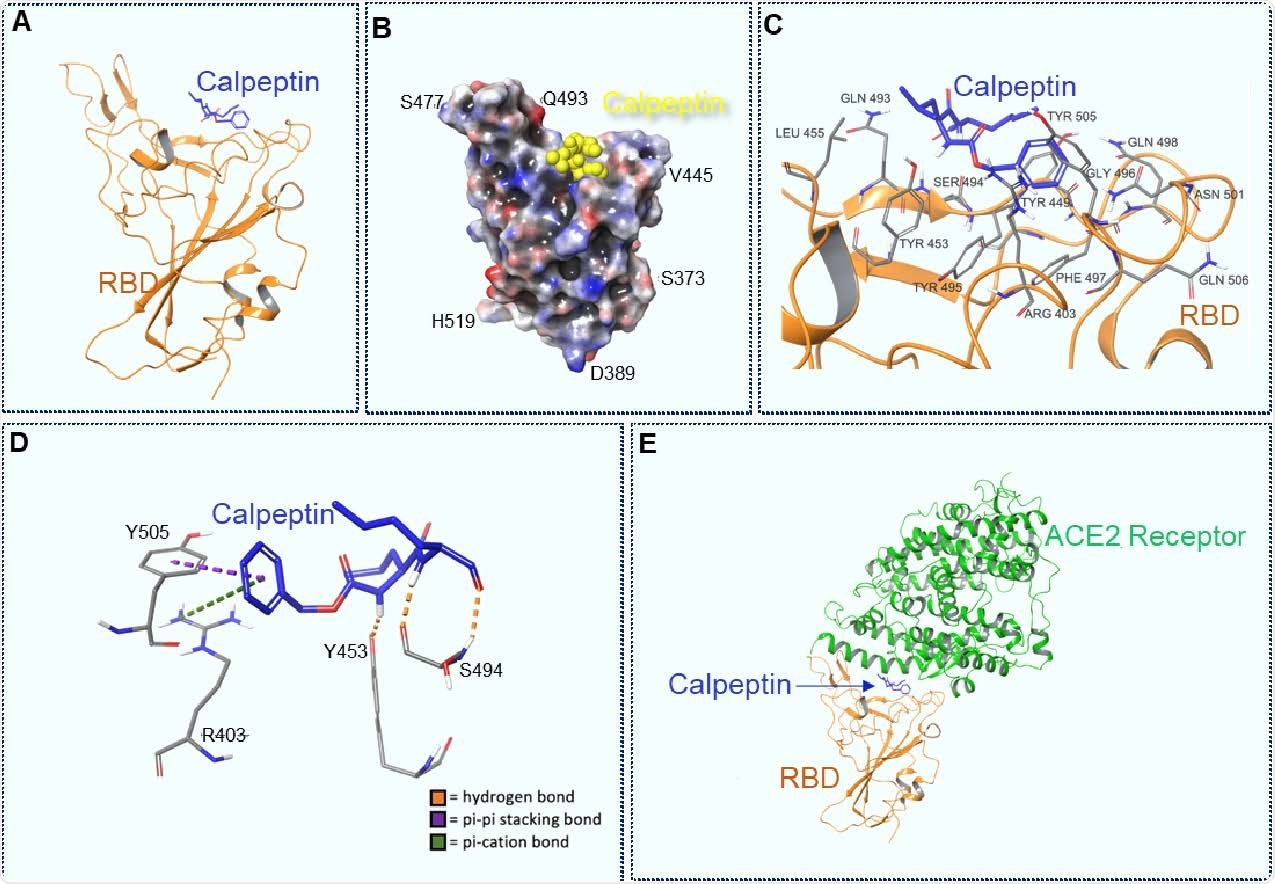
Docking of Calpeptin to the wild type RBD, using a modified PDB 6M0J structure. A. Calpeptin docked to the RBD. B. 3D view of Calpeptin docked to RBD. Molecular surface colored according to residue electrostatic potential. Calpeptin shown in yellow. C. Calpeptin’s interactions with critical residues within the RBD. D Types of interaction between Calpeptin and RBD. Hydrogen bonds are shown in orange, pi-pi stacking bons are shown in purple and pi-cation bonds are shown in green E. Calpeptin docked within the RBD-ACE2 connective interface.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Antiviral effects of calpeptin
As observed in the study, calpeptin significantly prevents the entry of wild-type SARS-CoV-2 into host cells by inhibiting both ACE2-mediated and endosomal entry pathways. A comparable inhibitory potency of calpeptin was also observed in post-entry steps.
Compared to the wild-type virus, calpeptin showed lower potency in inhibiting the entry of B.1.1.7 and B.1.351 variants. However, all inhibitions occurred at nanomolar concentrations.
To assess the specificity of calpeptin, the scientists conducted entry assays using ACE2-expressing cells that were infected with pseudotyped viruses containing SARS-CoV-2 spike or SARS-CoV spike. The findings revealed that calpeptin had an 8-fold lower efficacy against SARS-CoV than that against SARS-CoV-2.
Calpeptin-mediated inhibition of viral entry
The scientists conducted molecular docking of calpeptin to wild-type spike receptor-binding domain (RBD) in respect to ACE2. The findings revealed that calpeptin binds to wild-type RBD with the highest affinity by interacting with residues S494, Y453, Y505, and R403. Importantly, all these residues are vital for ACE2 – RBD interaction, and thus, for the viral entry. Among tested SARS-CoV-2 variants, calpeptin showed the lowest affinity for B.1.1.7 RBD. This could be because of the conformational changes induced by N501 spike mutation, resulting in a lesser number of intermolecular interactions between calpeptin and B.1.1.7 RBD.
Study significance
The study identifies calpeptin, a protease inhibitor, as a potent inhibitor of SARS-CoV-2 entry, with similar potency as the antiviral medicine remdesivir. The compound also demonstrates antiviral activity against SARS-CoV-2 variants, including B.1.1.7 and B.1.351. A high-affinity binding between calpeptin and spike RBD could be the primary mechanism of inhibiting RBD – ACE2 interaction and subsequently blocking the SARS-CoV-2 entry into host cells. However, inhibition of cathepsin L by calpeptin could be another potential mechanism of blocking viral entry at the cell surface.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Jablonski S. 2021. Identification of Potent Small Molecule Inhibitors of SARS-CoV-2 Entry, bioRxiv, https://doi.org/10.1101/2021.08.05.455262, https://www.biorxiv.org/content/10.1101/2021.08.05.455262v1
- Peer reviewed and published scientific report.
Mediouni, Sonia, Huihui Mou, Yuka Otsuka, Joseph Anthony Jablonski, Robert Scott Adcock, Lalit Batra, Dong-Hoon Chung, et al. 2022. “Identification of Potent Small Molecule Inhibitors of SARS-CoV-2 Entry.” SLAS Discovery 27 (1): 8–19. https://doi.org/10.1016/j.slasd.2021.10.012. https://slas-discovery.org/article/S2472-5552(21)00012-5/fulltext.