To date, the rapid spread of acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected more than 205 million individuals and claimed more than 4.3 million lives worldwide. In March 2020, the World Health Organization declared the coronavirus disease 2019 (COVID-19), caused by SARS-CoV-2, a pandemic. Even though a few drugs and vaccines are available to manage SARS-CoV-2 infection, they are not enough to bring the world back to normalcy and, therefore, there is an urgent need for more potent antiviral agents.
Background
SARS-CoV-2 is a positive-sense, single-stranded RNA virus belonging to the genus Betacoronavirus (β-CoV) of the family Coronaviridae. The viral genome contains 5’ untranslated region (UTR), structural genes, replicase complex (orf1ab), 3’ UTR, and accessory genes. The structural genes encode structural proteins, namely, spike (S), envelope (E), membrane (M), and nucleocapsid (N) protein. The S protein of the virus mediates the virus-host interaction such that it gains entry into the host cell. The S proteins contain two subunits, S1 and S2. Typically, the S protein is trimeric and contains a receptor-binding domain (RBD) on its S1 subunit, which is highly antigenic. S1 is responsible for the binding of the virus to the host receptor, while S2 mediates the fusion of membranes.
Why is it important to study the receptor-binding domain of the S protein?
Previous studies have reported that several neutralizing monoclonal antibodies (nmAbs) have been developed against the RBD of severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV). These nmAbs have potential application in therapeutics.
Several studies have also identified the crystalline structure of the RBD in complex, i.e., the complex structure formed after the binding of the SARS-CoV-2 S protein to human angiotensin I-converting enzyme 2 (hACE2). The hACE2-binding ridge in SARS-CoV-2 RBD binds to hACE2 more compactly than SARS-CoV RBD, hence why SARS-CoV-2 is more infectious than SARS-CoV. However, potent neutralizing antibodies with a higher affinity can inhibit the binding of RBD to hACE2.
Monoclonal antibodies - a powerful agent to combat infectious diseases
The mAbs, identified using RBD of SARS-CoV and MERS-CoV, can neutralize the virus directly by blocking the binding sites of the receptor on the RBD. In addition, these antibodies also induce a conformational change in the RBD that assists in preventing RBD binding to the receptor.
To date, researchers have identified and characterized many neutralizing antibodies obtained from transgenic mice and convalescent SARS-CoV-2 patients. Some of these antibodies have received emergency use authorization (EUA) from the Food and Drug Administration, USA, to treat SARS-CoV-2 infection.
Identification of neutralizing antibodies against SARS-CoV-2
A new study published in the journal Nature Communications has reported two neutralizing antibodies, namely, nCoVmab1 and nCoVmab2, which are potent against SARS-CoV-2.
Researchers have identified these antibodies from an extensive naïve human phage-displayed Fab library. They determined the specific epitopes of nCoVmab1 and nCoVmab2 by alanine scanning to understand the neutralizing mechanism.
The researchers of this study have also examined the binding abilities using the germline formats of nCoVmab1 and nCoVmab2 to the RBD, helping to reveal the significance of the antibody maturation pathway, which is responsible for eliciting potent neutralizing antibodies by RBD.
The authors of this study reported that both the antibodies, nCoVmab1 and nCoVmab2, showed high neutralizing ability against pseudotyped SARS-CoV-2, as well as pseudotyped SARS-CoV-2 D614G. However, between the two, nCoVmab1 showed better neutralizing capability against SARS-CoV-2’s original strain by inhibiting the receptor from binding to host cells.
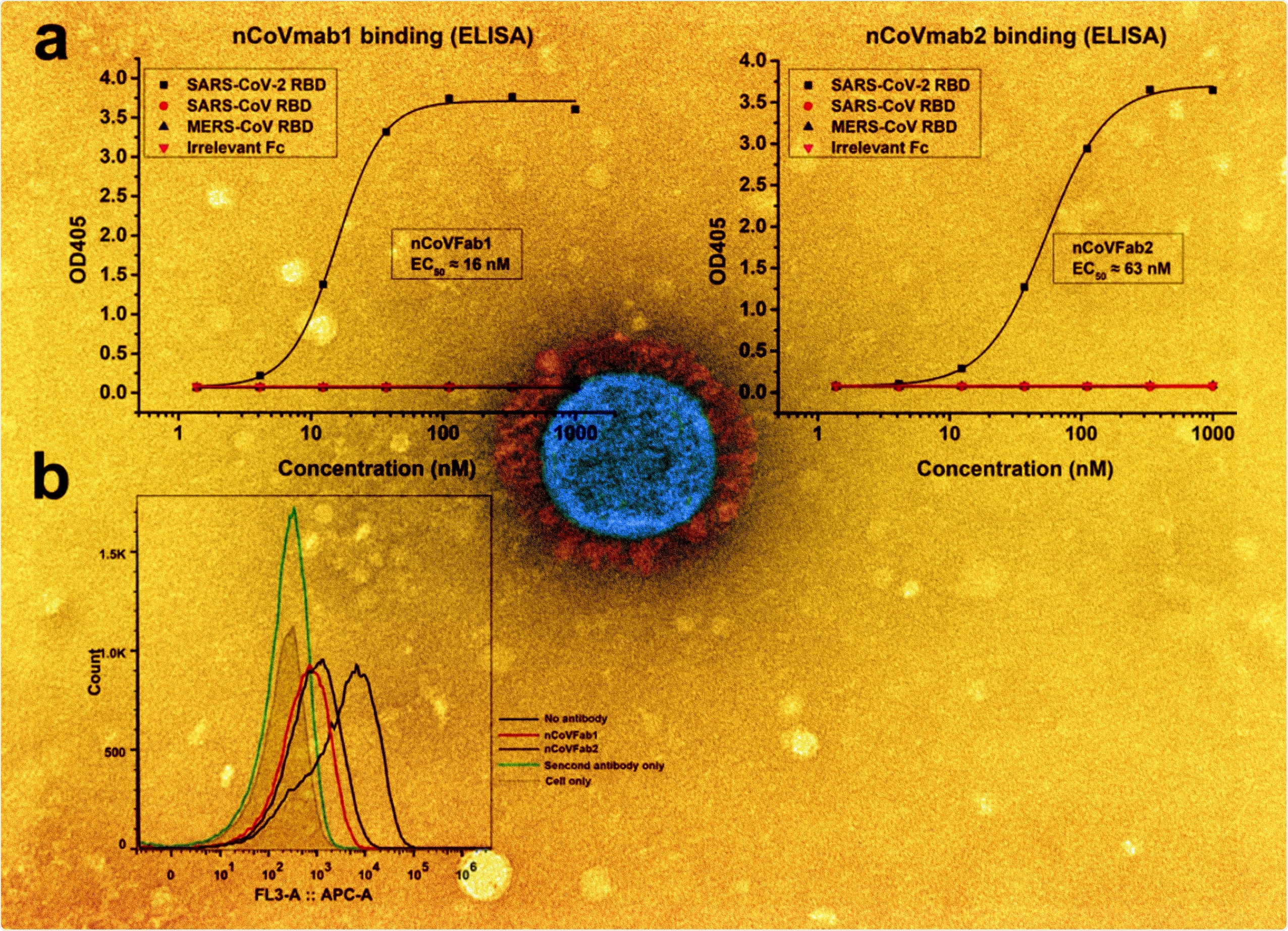
a Binding of nCoVFab1 and nCoVFab2 to different RBDs, as measured by an ELISA. RBD-Fc fusions of SARS-CoV-2, SARS-CoV, and MERS-CoV were used as antigens. Irrelevant Fc protein was used as a negative control. The experiments were performed independently twice and similar results were obtained. One representative experiment is shown and data are average values of two replicates. b Blockage of binding of SARS-CoV-2 RBD-Fc to Vero E6 cells by Fabs. nCoVFab1 and nCoVFab2 inhibited the binding fluorescence shift with efficiencies of approximately 89% and 82%, respectively, as measured by flow cytometry.
The researchers revealed that both the antibodies could prevent virus plaque spread and that the cytopathic effect (CPE) was observed. They further reported no issues with the stability, autoreactivity, or significant drawbacks of the antibodies.
The scientists further determined the efficacy of nCoVmab1 using the K18-hACE2 transgenic mouse model. This study found that nCoVmab1 could significantly reduce lung viral titers by 10-10000-fold and provide prominent protection to lung lesions. Thereby, nCoVmab1 was found to possess a promising prophylactic and therapeutic capability.
Both nCoVmab1 and nCoVmab2 are germline-like antibodies that competitively interact with RBD of the SARS-CoV-2’s S protein and successfully prevent the binding between RBD and hACE2.
Previous studies reported that typically germline-like antibodies contain fewer mutations compared to the corresponding matured antibody. Further, in the current study, the alanine scanning revealed the amino acids involved in RBD’s binding to hACE2. This helped researchers to understand the mechanism by which the virus evades immune surveillance by neutralizing antibodies. It also helped select antigenic targets for the development of potent antiviral agents against SARS-CoV-2 variants.
Conclusion
Even though both nCoVmab1 and nCoVmab2 have similar heavy chains, they have entirely different light chains. This difference may be the reason for their differential affinity and neutralizing activity. The somatic hypermutation rate of the heavy chain is very low, and nCoVmab1 possesses more SHMs in its light chain.
The authors of this study have stated that nCoVmab1 is much more potent than nCoVmab2 and could be used as a potential therapeutic agent for fighting SARS-CoV-2 infection.