Scientists have stated that molecular mimicry between human proteins and pathogens can incorrectly lead to antibodies attacking human proteins. Such an occurrence can cause transient or chronic autoimmune disorders.
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infected individuals often suffer from thrombocytopenia, a condition associated with low platelet counts. The cytokine thrombopoietin regulates platelet count. Previous studies have reported that thrombocytopenia can increase the coronavirus disease 2019 (COVID-19) mortality rate by about five times.
Thrombocytopenia and COVID-19
Researchers revealed that thrombocytopenia in COVID-19 patients is similar to immune thrombocytopenia, which is why autoantibodies mistakenly target human thrombopoietin (hTPO) and its receptor, leading to a decrease in platelet count. For those patients with and without COVID-19, the researchers found that treatments with TPO Receptor Agonists improve thrombocytopenia, showing that mistaken targeting occurs before TPO activates the TPO receptor.
Scientists searched for a molecular resemblance between the spike protein of SARS-CoV-2 and human epitopes from The Immune Epitope Database (IEDB). A five-amino acid motif, namely, TQLPP, was identified that shared structural similarity with the Spike and hTPO proteins. In the spike protein, the motif is present on the surface of the N-terminal Domain (NTD), while in the case of the hTPO, the motif is present at the interface with a neutralizing antibody.
A New Study
A new study published on the bioRxiv* preprint server hypothesizes that SARS-CoV-2 infection may induce the production of TQLPP-specific antibodies against its epitope that can cross-react with hTPO. The present research uses molecular modeling and machine learning techniques to investigate the binding of an hTPO neutralizing mouse Fab antibody (TN1) to the Spike TQLPP epitope in the absence of Spike TQLPP antibodies.
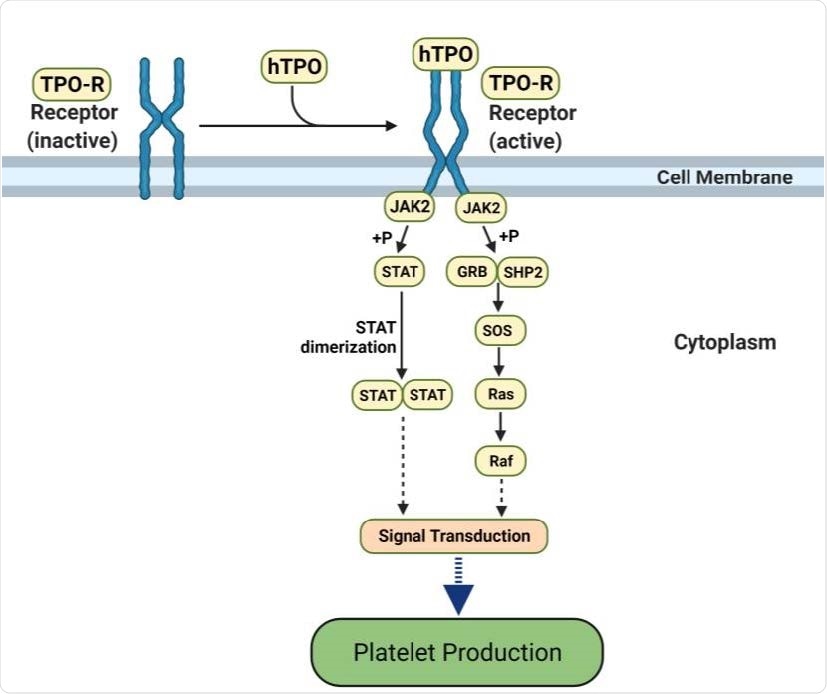
hTPO pathway to induce platelet production. Simplified JAK-STAT signaling pathway in megakaryocytes where hTPO activates the TPO receptor and triggers signaling cascades that stimulate platelet production (44, 45). Created with BioRender.com.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The present research constructs a composite model of the Spike protein and TN1 by coupling a structure of hTPO complexed with TN1 Fab with a published full-length glycosylated model of the Spike protein. For this research, the first 26 residues that contained the TQLPP motif were used from the glycosylated model based on PDB id 6VSB. Using molecular dynamics (MD) simulations, researchers minimized the energy and equilibrated the Spike-TN1 complex.
In this model, researchers found that the Spike trimer formed a complex with three TN1 Fab antibodies. The TQLPP epitope was found to be available to the antibody without encountering any inhibitions from neighboring glycan chains. Generally, adjacent glycans shield the antibody-binding site. In this context, researchers calculated root mean square deviation (RMSD) for TQLPP regions using 60 Spike proteins from Protein Data Bank (PDB) to confirm the conformation of TQLPP.
The MD simulation, associated with hTPO and Spike NTD with TQLPP complexed with the TN1 antibody, helped evaluate the molecular mimicry between the antibody interface areas. Researchers further substantiated their model’s findings by evaluating the antibody-antigen interface complementarity with MaSIF-search.
MaSIF-search is a recent tool, which is used to calculate the binding score. The binding score correlates with the strength of binding when forming a stable complex. A Lower score represents stronger binding. The results obtained using this tool showed Spike-TN1 complexes have lower binding scores than random complexes.
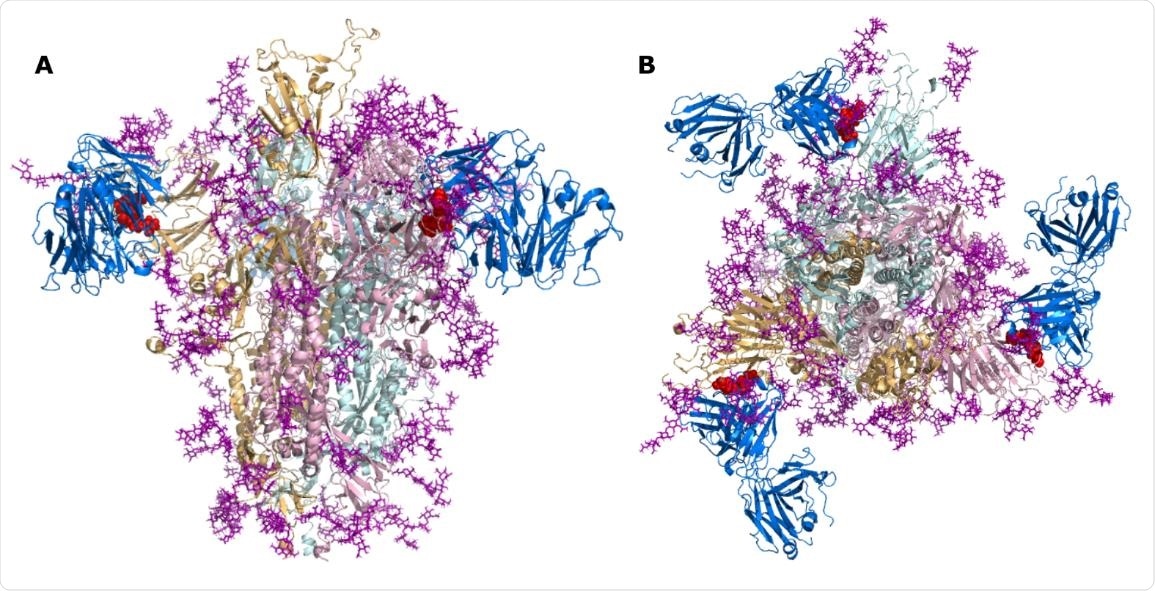
SARS-CoV-2 Spike bound to TN1 Fab antibody. SARS-CoV-2 Spike shown in the trimeric state (PDB id: 6VSB) bound to TN1 Fab antibody (blue, PDB id: 1V7M) as viewed from (A) the side and (B) the top. The TQLPP motifs are shown as red spheres and glycans are shown in purple.
To determine structural mimicry between Spike-TQLPP and all human-TQLPP motifs, the authors of this study utilized AlphaFold prediction. The closest structural mimic was hTPO, followed by coiled-coil domain-containing protein 185, Fc receptor-like protein 4 (FCRL4), and far upstream element-binding protein 1. These predictions prove that TQLPP motifs have similar conformations, thereby supporting the concept of structural mimicry.
The potential cross-reactivity of an antibody targeting TQLPP in the proteins mentioned above was studied. Researchers eliminated six proteins that exhibited the TQLPP motif in low confidence or unstructured regions, and three proteins, namely, NEK10 (ciliated cell-specific kinase), FCRL4, and ALG12, were complexed with TN1. Among these three protein complexes, the binding score of NEK10-TN1 was analogous to the hTPO-TN1 complex. Previous studies revealed that NEK10 controls the motile ciliary function, leading to eliminating pathogens from the respiratory tract. Therefore, NEK10 dysfunction can hinder mucociliary clearance, resulting in respiratory disorders, e.g., bronchiectasis.
Conclusion
The current research has indicated a high possibility of cross-reactivity between the Spike protein of SARS-CoV-2 and hTPO involving the TQLPP epitope. This event affects the production of platelets leading to thrombocytopenia. Additionally, the cross-reactivity with other TQLPP-containing proteins, e.g., NEK10, also supports the hypothesis. Also, researchers reported that the presence of neutralizing antibodies against peptides with TQLPP in the plasma of the COVID-19 recovered patients, especially in severe cases. Which further adds credence to the research hypothesis. Notably, the COVID-19 vaccines that are developed based on Spike protein of SARS-CoV-2 can cause thrombocytopenia.
The authors of the current study propose that scientists should consider avoiding potential predictable autoimmune interference for the future development of the COVID-19 vaccines. Another interesting point to note is the evolutionary trend in the TQLPP motif, i.e., it might not remain in the Spike region. Such an occurrence has been observed in the SARS-CoV-2 Gamma variant. The neutralizing antibodies targeting the NTD supersite may become ineffective against the SARS-CoV-2 variant due to rapid mutation. However, for future COVID-19 vaccines, protein engineering of the TQLPP motif may reduce the possibility of thrombocytopenia and provide long-term protection against future SARS-CoV-2 variants.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Nunez-Castilla, J., et al. (2021) Molecular mimicry between spike and human thrombopoietin may induce thrombocytopenia in COVID-19. bioRxiv 2021.08.10.455737; doi: https://doi.org/10.1101/2021.08.10.455737, https://www.biorxiv.org/content/10.1101/2021.08.10.455737v1
- Peer reviewed and published scientific report.
Nunez-Castilla, Janelle, Vitalii Stebliankin, Prabin Baral, Christian A. Balbin, Masrur Sobhan, Trevor Cickovski, Ananda Mohan Mondal, et al. 2022. “Potential Autoimmunity Resulting from Molecular Mimicry between SARS-CoV-2 Spike and Human Proteins.” Viruses 14 (7): 1415. https://doi.org/10.3390/v14071415. https://www.mdpi.com/1999-4915/14/7/1415.