Scientists predict the COVID-19 pandemic is on its way to becoming endemic. The rising spread of COVID-19 variants such as Delta has helped infect unvaccinated and some vaccinated individuals. COVID-19 vaccines continue to be the best way to protect against severe illness and death, but there may be a need for vaccines to protect against a specific variant.
Research led by Vivek R. Nerurkar from the University of Hawaiʻi at Mānoa developed a mathematical algorithm that can help optimize the vaccine platform. The algorithm monitors the evolution of SARS-CoV-2 lineages and incorporates information such as spike mutations from variants of concern, SARS-CoV-2 genomes, and clinical samples.
The researchers write:
Our findings have relevance to the future of tracking SARS-CoV-2 and of SARS-CoV-2 vaccine design…If established in real-time, the herein described algorithm will allow researchers to understand the evolving SARS-CoV-2 genome preemptively rather than responsively.”
The study “Algorithm for the Quantitation of Variants of Concern for Rationally Designed Vaccines Based on the Isolation of SARS-CoV-2 Hawaiʻi Lineage B.1.243” is published on the bioRxiv* preprint server, while awaiting peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Validating the algorithm using SARS-CoV-2 isolates from the B.1.243 lineage
The researchers applied an adaptive algorithm to monitor SARS-CoV-2 evolution and other genetic changes. The goal was to use the information to help design logical next-generation vaccines.
They collected SARS-CoV-2 samples 5 days after two patients tested positive for SARS-CoV-2. Genomic sequencing was performed and compared to data from both GenBank and GISAID to identify the SARS-CoV-2 lineage. The samples were found to be related to the B.1.243 lineage.
The B.1.243 variant once made up 72% of genomic sequences from Hawaii. However, by July 28, 2021, the variant decreased to about 23% of all sequences. At the time of the study, the Delta variant was taking over as the dominant SARS-CoV-2 strain in the area.
The SARS-CoV-2 genome evaluation showed 49 mutations in the SARS-CoV-2 isolates related to the B.1.243 lineage. About 9 of the spike protein mutations observed had a 70% prevalence worldwide. The 10th spike protein mutation was D614G, which is widespread in more than 99% of strains.
Isolating SARS-CoV-2 from samples and analyzing published sequences showed that B.1.243 made up more than 40% of all cases in Hawaii. However, the researchers note that it is unlikely to be considered a variant of concern given its decreasing prevalence worldwide.
B.1.243 helped validate the algorithm and analyze variants of concern/interest based on emerging spike protein amino acid changes for future surveillance and vaccine design.
The algorithm provided numerical value to each variant of concern and predicted the likelihood it would spread across the population. It also calculated the probability each variant of concern would undertake future mutations.
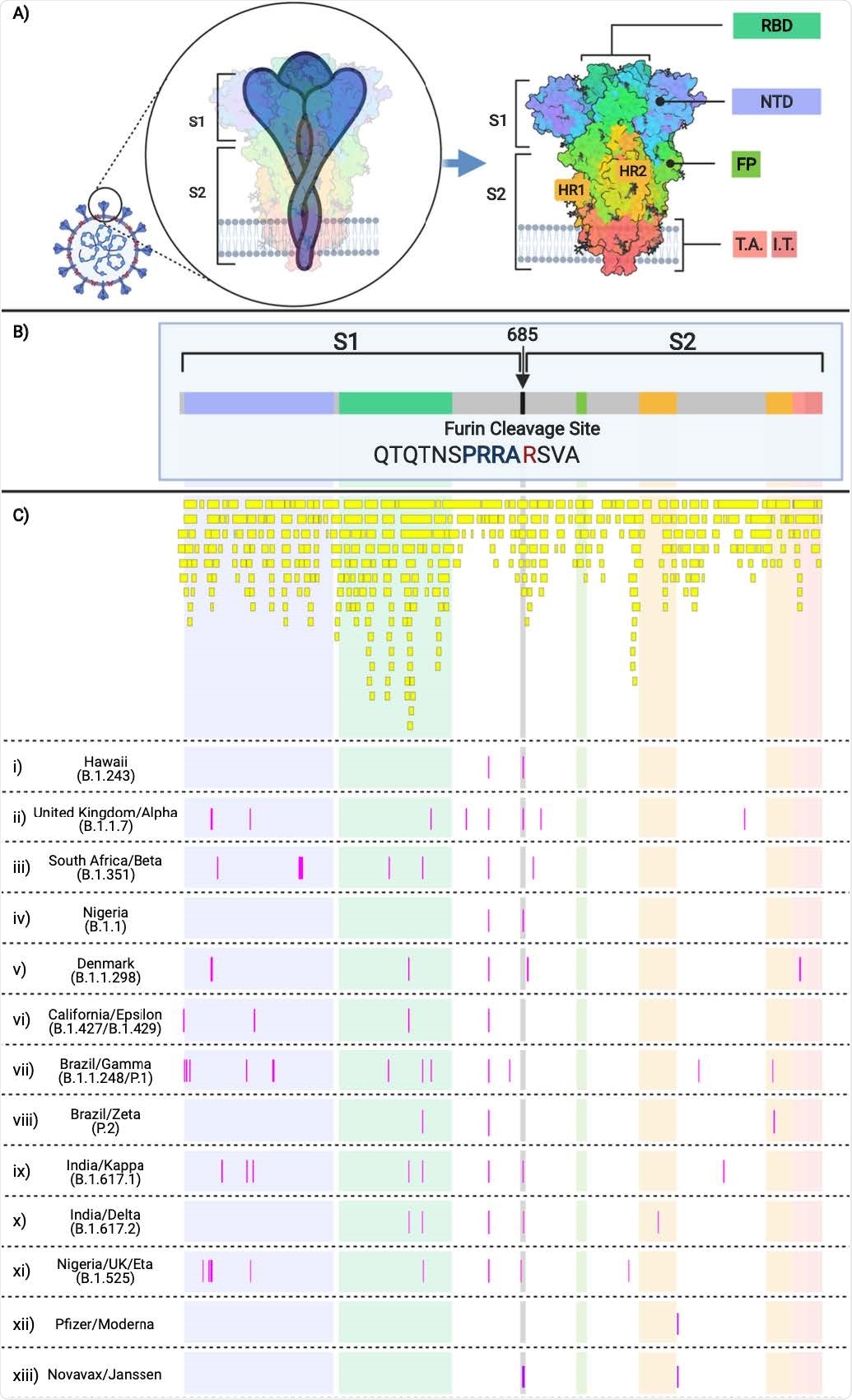
SARS-CoV-2 Spike Protein Domains and Relation to B and T cell Epitopes, Variant Amino Acid Substitutions, and Vaccine Amino Acid SubstitutionsThis figure demonstrates the evolution of the SARS-CoV-2 variants by depicting the location of the variants substitutions and deletions in the context of spike domains and epitopes. A) Cartoon rendering of SARS-CoV-2 and the 1,273 amino acid long spike protein overlay onto the color-coded crystallographic structure determined by electron microscopy (PBD ID: 6VXX-PDB). The individual protein domains are color-coded: N-terminal domain (NTD) (light purple) (residues 14-305), receptor-binding domain (RBD) (teal green) (residues 319-541), furin (F) (residues 682-685), fusion protein (FP) (green) (residues 788-806), heptad repeat 1 (HR1) (orange) (residues 912-984), heptad repeat 2 (HR2) (orange) (residues 1163-1213), transmembrane anchor (TM) (light pink) (1213-1237), and intracellular tail domain (IT) (dark pink) (1237-1273). B) Two-dimensional layout of the spike protein and domains with the addition of the S1/S2 furin cleavage site (RRA/R) (682-685) (black). C) In silico predicted B and T cell epitope loci revealing 393 in silico B and T cell epitopes mapped here individually as a yellow boxes i)-xiii) Amino acid substitutions present in the corresponding variant shown in pink boxes in comparison to the reference sequence NC_045512. i) B.1.243 Hawaii; ii) B.1.1.7 United Kingdom; iii) B.1.351 South Africa; iv) B.1.1 Nigeria; v) B.1.1.298 Denmark; vi) B.1.427 and B.1.429 California; vii) P.1 Brazil; viii) P.2 Brazil; ix) B.1.617.1 India; x) B.1.617.2 India; xi) B.1.525 United Kingdom/Nigeria; xii) Pfizer and Moderna mRNA sequences with artificially added substitutions K986P and V987P; xiii) Novavax and Janssen mRNA sequences with artificially added substitutions R682S/Q, R683Q, R685G/Q, K986P, and V987P.
Spread of variant of concerns, especially Gamma, are projected to increase worldwide
The research team used the data to predict exponentially emerging mutation to the SARS-CoV-2 isolates in the current study.
The Centers for Disease Control and Prevention currently identifies Alpha, Beta, Delta, and Gamma as four variants of concern.
Results suggest all four variants of concern will have an exponential spread, with Gamma being the most dominant worldwide.
“The Delta variant, predicted to be exponentially emerging by this quantitative analysis with an r-value of 0.96 as of April 2021 at 1% prevalence, has become the most prevalent worldwide as of June 2021, representing 64% of worldwide sequences,” wrote the team.
Based on the findings, the team suggests the algorithm can serve to monitor for SARS-CoV-2 mutations and serve as a baseline for choosing the primary structure of vaccines. They also argue that it’s a valuable tool in taking a proactive and predictive approach to dealing with future variants.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Maison DP, et al. Algorithm for the Quantitation of Variants of Concern for Rationally Designed Vaccines Based on the Isolation of SARS-CoV-2 Hawaiʻi Lineage B.1.243. medRxiv, 2021. doi: https://doi.org/10.1101/2021.08.18.455536, https://www.biorxiv.org/content/10.1101/2021.08.18.455536v1
- Peer reviewed and published scientific report.
Maison, David P., Lauren L. Ching, Sean B. Cleveland, Alanna C. Tseng, Eileen Nakano, Cecilia M. Shikuma, and Vivek R. Nerurkar. 2022. “Dynamic SARS-CoV-2 Emergence Algorithm for Rationally-Designed Logical Next-Generation Vaccines.” Communications Biology 5 (1): 1–12. https://doi.org/10.1038/s42003-022-04030-3. https://www.nature.com/articles/s42003-022-04030-3.