The coronavirus disease (COVID-19) pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), threatens global public health with total cases and deaths reaching over 212 million and 4.44 million, respectively.
Though vaccination efforts are underway in most countries, there are still limited options for treating COVID-19. Now, researchers at Penn State designed a new COVID-19 therapy that uses a defective synthetic version of the SARS-CoV-2 virus to interfere with the actual virus's replication.
Published in the journal PeerJ Life & Environment, the study highlights the use of the defective virus that replicates three times faster due to its shorter size, interfering with the replication of the real virus. The defective synthetic version could be used as a self-promoting antiviral therapy, wherein the synthetic version replicates faster, the virus will assist in its own demise.
Defective genome replication
Versions of a viral genome with large deletions frequently emerge from most ribonucleic acid (RNA) viruses. Defective genomes lacking essential coding sequences can still replicate and packaged into virions in the presence of active viruses.
The full viral genome produces the essential proteins for replication, which can be exploited by defective genomes that retain the ability to bind to these proteins. Hence, these defective genomes are considered parasites of the full-length virus since they compete for replication. Since they are shorter in length, they can replicate faster than their full-length parental genome in cells.
Common coronaviruses may contain these genomes, called defective interfering (DI) genomes. In SARS-CoV-2, long deletions have been reported, and DI genomes have been shown to emerge by recombination caused by sequence microhomology.
The study
In the study, the researchers created short synthetic DI RNAs from parts of the wild-type SARS-CoV-2 genome to examine whether they could replicate in coinfected cells and be packaged into virions. They quantified the relative amounts of the DI and WT genomes in the cells over periods, showing the interference of the DI genome with the wild-type genome (WT).
Synthetic defective interfering viruses. (A) Three portions of the wild type (WT) SARS-CoV-2 genome were used to create a synthetic defective interfering genome (DI1) and a shorter version (DI0) comprising only parts of the two terminal portions. Numbers delimiting the portions refer to positions in the SARS-CoV-2 genome. The first position is mutated (A →C) in both DI1 and DI0. Open rectangles show the position of the probes and primers used. (B) To produce synthetic DI particles, DNA constructs corresponding to the RNA sequence of DI1 or DI0 were transcribed into RNA in vitro using T7 RNA polymerase and transfected into Vero-E6 cells that were then infected with SARS-CoV-2. The supernatant from these cell cultures was used to infect new cells.
The study results showed that the defective synthetic genome replicates three times faster than SARS-CoV-2 in coinfected cells and interferes with it, reducing the viral load by about half in 24 hours. No differences between the packaging efficiencies of the two genomes were found, as transmission rates were the same. Hence, it can be concluded that the reduced amount of WT genomes was due to interference as a result of the faster replication of the DI genome. Significantly, amounts of DI genome so small they are undetectable via qRT-PCR can interfere with the WT virus.
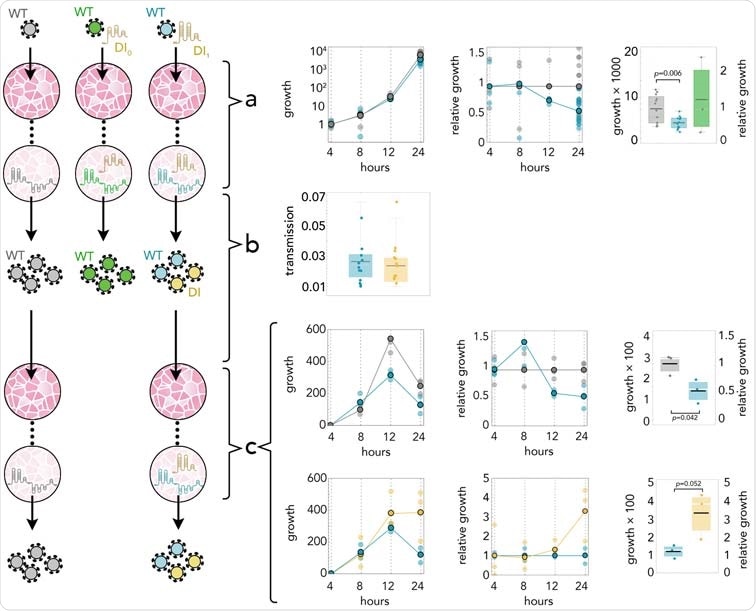
DI1 reduces the amount of SARS-CoV-2 by half; it replicates 3 times faster; and it is transmit- ted with the same efficiency. (A) Growth rates (absolute amount relative to the amount at 4 h) of WT in controls (gray) and in coinfections with DI1 (blue) or DI0 (green); growth relative to controls at the same time point; and detail at 24 h. (B) Transmission efficiency of WT (blue) and DI1 (yellow) in coinfections: the amount, measured by qRT-PCR, immediately before passaging divided by the average amount mea- sured almost immediately (4 h) after passaging (using the supernatant to infect new cells 24 h after initial infection). DI0 was detected inside the cells but not in the supernatant. (C) Growth rates (absolute amount relative to the amount at 4 h) of WT in controls (gray) and in coinfections (blue); growth relative to con- trols at the same time point; and detail at 24 h. Growth rates (absolute amount relative to the amount at 4 h) of WT (blue) and DI1 (yellow) in coinfections; growth relative to that of WT in coinfections at the same time point; and detail at 24 h.
Indeed, DIs could be used as antivirals since they replicate faster in cells and interfere with the real virus. Meanwhile, the team explained that as the DI genomes increase in frequency among the virus particles pool, the process becomes more effective until the decline in the amount of the wild type SARS-CoV-2 leads to the demise of both the virus and DI. A similar approach can be used in bacterial infections and cancer.
"We have established a proof of principle that a synthetic defective interfering SARS-CoV-2 can replicate in cells infected with the virus and interfere with its replication," the researchers concluded in the study.
The team added that further studies and experiments are necessary to verify the potential of SARS-CoV-2 Dis as an antiviral treatment. These studies need to determine the method's efficacy in human lung cell lines and against some of the newer SARS-CoV-2 variants of concern (VOC). It would also be useful to analyze the evolution of coinfections long-term to test how SARS-CoV-2 and its DIs coevolve and, importantly, investigate whether resistant mutants can arise.
Source:
Journal reference: