Genomic surveillance is essential before, during, and after an infectious outbreak to guide and implement public health interventions. However, there are striking differences in the spatial and temporal intensity of genomic surveillance worldwide.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
COVID-19 is caused by an RNA virus, the severe acute respiratory syndrome coronavirus 2 (SARS-COV-2). RNA viruses undergo high mutations and accumulate genetic changes at high evolutionary rates. These changes allow the virus to adapt to selective pressures induced by antivirals, vaccines, and host immunity, leading to the emergence of viral variants.
Since its outbreak in December 2019, the SARS-CoV-2 has emerged as many new variants that pose an increased risk to public health. The variants of concern (VOCs) (such as Alpha/B.1.1.7; Beta/B.1.351; Gamma/P.1; and Delta/B.1.617.2 (and its descendent AY lineages) have genotypic and phenotypic traits that may affect the diagnostics or therapeutics, confer higher transmissibility, lead to higher disease severity, and allow immune escape from the natural infections or the vaccines. In addition, the variants of interest (VOI) (including Eta/B.1.525; Iota/B.1.526; Kappa/B.1.617.1 and; Lambda/C.37) share some genetic traits with VOCs and, therefore, pose similar possible risks.
The SARS-CoV-2 genetic diversity tracked in real-time allows identifying striking differences in the temporal and spatial spread of the virus worldwide.
A team of biologists, epidemiologists, and biomathematicians, looked at the disparities in genomic surveillance during the first 15 months of the COVID-19 pandemic.
Genomic surveillance and results from the study
The study reported that following the evolution of VOCs/VOIs, many countries have initiated or scaled up genomic surveillance, leading to an unprecedented number of viral genomes in publicly accessible databases, with >2,400,000 consensus genome sequences deposited in GISAID, >916,000 high-throughput sequencing datasets, and >969,500 consensus sequences in NCBI as of July 20th, 2021.
However, of the submitted genomes, 94% come from high-income countries (HIC) and only 6% from low and middle-income countries (LMICs). Out of the 167 countries studied, 100 countries sequenced <0.5% of confirmed cases, and only 16 countries were able to sequence >5% of their overall confirmed cases. Despite reporting a similar number of cases in the HICs and LMICs (65.3 and 61.2 million, respectively), they respectively sequenced 1.81% and 0.11% of their cases.
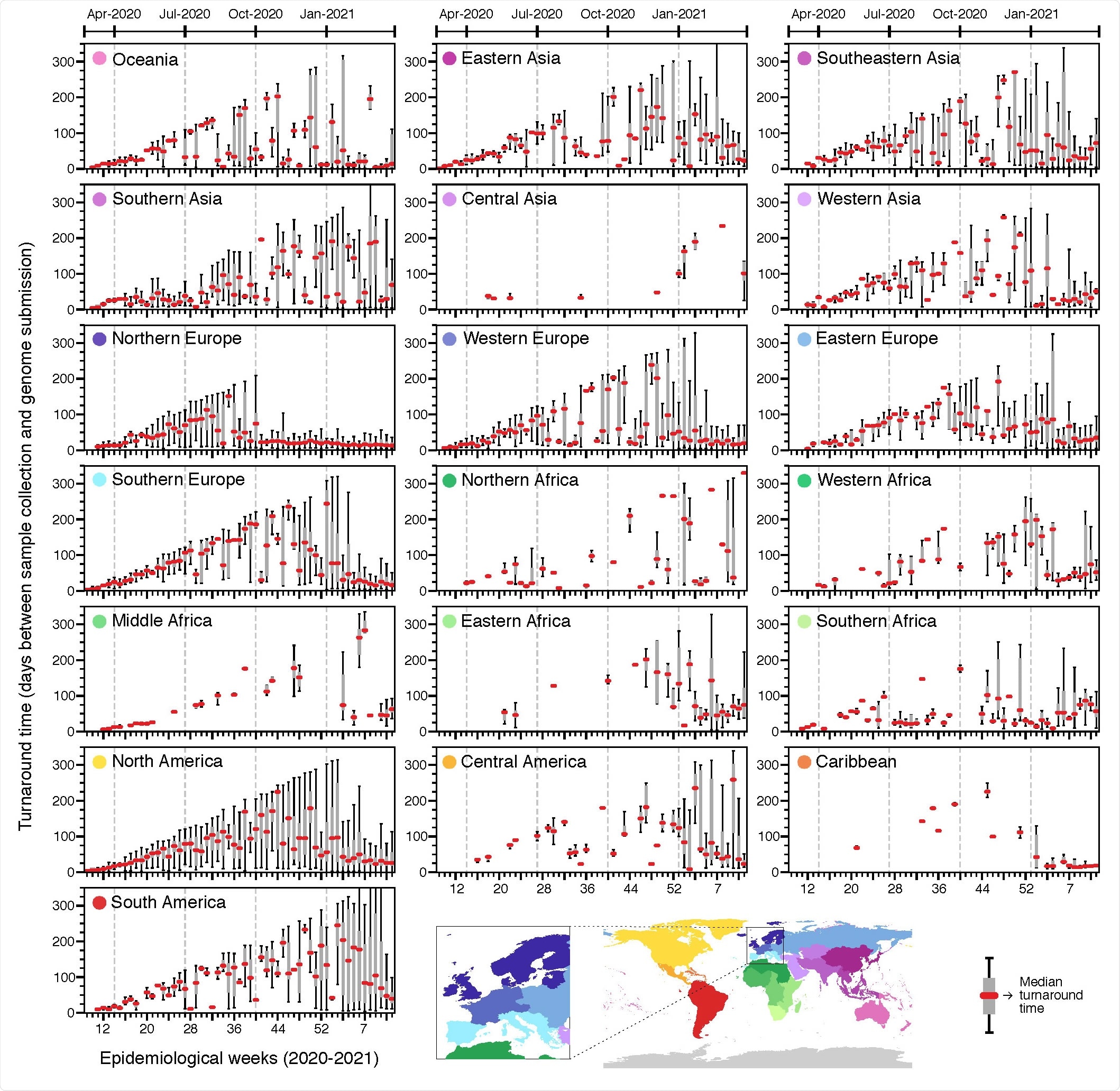
Turnaround time across geographic regions. Delays between sample collection and genome submission across epidemiological weeks (turnaround time) in different regions, between February 23rd, 2020 and March 27th, 2021, based on metadata submitted to GISAID up to May 30th, 2021.
The researchers found a moderate negative correlation between weekly sequencing percentages and the reported COVID-19 incidence. Countries such as Hong Kong, Taiwan, New Zealand, Australia, and Iceland that kept COVID-19 incidence at low levels generally could sequence a high proportion of cases.
Analyzing the weekly incidence and the sequencing rates, it was found that for most LMICs it is not feasible to sequence high or even moderate percentages of cases (0.1% to 1%) each week. Many do not have openly available genomes or are only represented in the global genomic surveillance because of cases associated with travel from those locations and being sequenced abroad.
On the other hand, European countries sequenced high or very high percentages of cases, nearly on a weekly basis.
“Since the initial detection and emergence of the VOC B.1.1.7/Alpha in the UK, countries across the world have sought to intensify genomic surveillance.”
Rapid public sharing of data is essential for genomic surveillance which varied greatly across geographic regions. Longer turnaround times (between sample collection and genome submissions) could result from any of the following reasons: to investigate reinfections, vaccine escape, to understand past epidemic dynamics, types of research that are slower than public health surveillance, insufficient lab personnel, delays in shipment of samples and reagents, and poor coordination and lack of experienced professionals.
To investigate the impact of socioeconomic factors on the SARS-CoV-2 genomic surveillance preparedness around the world, the researchers explored a list of country-level covariates and its correlation of the percentage of sequenced COVID-19 cases. These included expenditure on R&D per capita, GDP per capita, socio-demographic index, established influenza genomic surveillance capacity, and healthcare expenditure. These socioeconomic factors represent important obstacles and dictate the need for efforts to improve the genomic capacity in LMICs. This will prevent the unnoticed emergence and spread of variants.
“To start, diagnostic capacity needs to be enhanced, as case underreporting directly impacts the ability of countries to detect variants and their frequency changes.”
Based on the analysis in this study, the researchers proposed a 0.5% threshold could be achieved by sequencing 1 genome for every 200,000 habitants as a reasonable benchmark.
“Based on empirical data and our statistical analysis, if public health labs worldwide use such a benchmark to set their minimal operational limits to sequence at least 0.5% of the cases at high incidence (100 cases/100,000 pop.), with quick turnaround time (<21 days), it would greatly improve our global capacity to detect new variants and track changes in variant prevalence.”
In conclusion, this study provides a comprehensive overview of SARS-CoV-2 genomic surveillance patterns observed worldwide, highlighting disparities in surveillance capacity in different geographic regions regarding the percentage of sequenced cases, frequency of sampling, and turnaround time.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Global disparities in SARS-CoV-2 genomic surveillance, Anderson F. Brito, Elizaveta Semenova, Gytis Dudas, Gabriel W. Hassler, Chaney C. Kalinich, Moritz U.G. Kraemer, Sarah C. Hill, Danish Covid-19 Genome Consortium, Ester C. Sabino, Oliver G. Pybus, Christopher Dye, Samir Bhatt, Seth Flaxamn, Marc A. Suchard, Nathan D. Grubaugh, Guy Baele, Nuno R. Faria, medRxiv, 2021.08.21.21262393; doi: https://doi.org/10.1101/2021.08.21.21262393, https://www.medrxiv.org/content/10.1101/2021.08.21.21262393v1
- Peer reviewed and published scientific report.
Brito, Anderson F., Elizaveta Semenova, Gytis Dudas, Gabriel W. Hassler, Chaney C. Kalinich, Moritz U. G. Kraemer, Joses Ho, et al. 2022. “Global Disparities in SARS-CoV-2 Genomic Surveillance.” Nature Communications 13 (1): 7003. https://doi.org/10.1038/s41467-022-33713-y. https://www.nature.com/articles/s41467-022-33713-y.