The emergence of escape mutations in the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is somewhat less likely for the Delta variant in comparison with the original or wild type virus, shows a recent study currently available on the bioRxiv* preprint server while it undergoes peer review.
One of the most pressing issues during the coronavirus disease (COVID-19) pandemic, caused by SARS-CoV-2, is a potential emergence of viral escape variants that may render our current vaccines ineffective. This is especially concerning as the majority of the global population is still unvaccinated.
The interface between the receptor-binding domain (RBD) of the SARS-CoV-2 spike glycoprotein and the receptor on host cells known as angiotensin-converting enzyme 2 (ACE2) mostly overlaps with binding sites for the most potent neutralizing antibodies we have, limiting the repertoire of likely mutations.
Consequently, mutations arising in the RBD may subtly change the interface between RBD and ACE2, but also between RBD and neutralizing antibodies. Furthermore, these mutations may interact (which is known as epistatic interactions) and subsequently increase the risk of vaccine escape.
And while the structures of the RBD-ACE2 interface in wild-type SARS-CoV-2 are quite similar to that of the RBD-NAb interface, just one mutation in the RBD can induce distinct shape changes in both interfaces and present with an additive effect.
A research group led by Dr. Nash D. Rochman from the National Center for Biotechnology Information, National Library of Medicine in Bethesda (United States) used unique protein structure modeling in order to study the potential of escape mutations.
Predicting the effects of mutations
In short, this study used an expensive computational approach to predict the effects of all single mutants at the RBD-NAb and RBD-ACE2 interfaces for the wild type SARS-CoV-2, but also its Gamma variant and Delta variant on receptor and antibody binding.
An ensemble of 50 native conformations per complex has been approximated with the use of standard Rosetta protocols. More specifically, after charting the wild-type SARS-CoV-2 RBD landscape, the researchers aimed to discern the most prominent combinations of RBD mutations that were in circulation over the course of the COVID-19 pandemic.
Moreover, the positive cost of the increase and the negative cost of the decrease in the ACE2 or neutralizing antibody binding affinity relative to the wild-type virus has been assessed in order to see whether it provides a selective advantage.
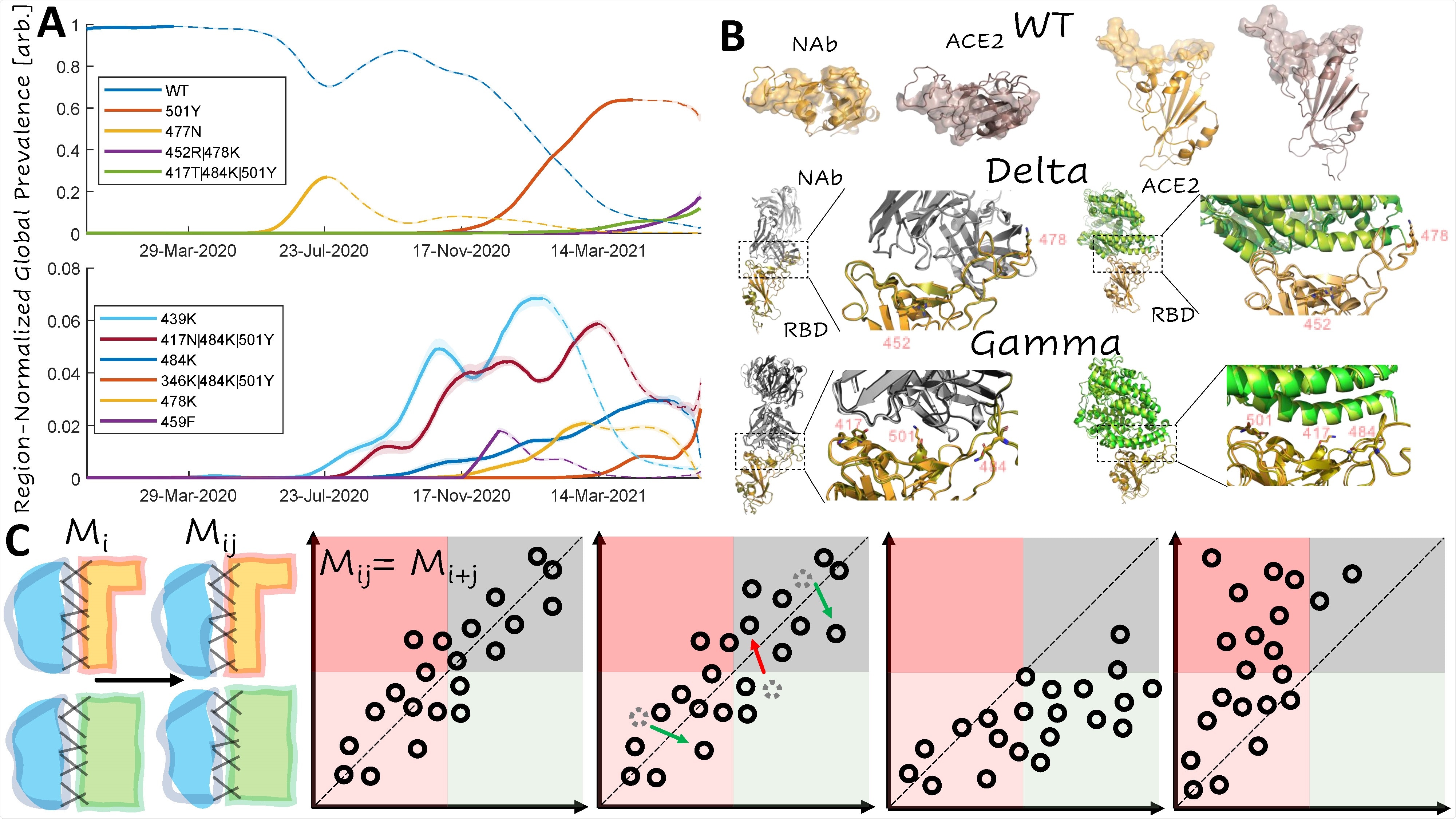
Dominant Trends in Circulating RBD Mutations A. Region-normalized global prevalence of the top 10 most common combinations of RBD mutations over time. Lines are solid up to peak prevalence and dashed afterwards. Shading indicates confidence intervals. B. Structural comparison of the complexes of ACE2 and NAb with RBD for WT, Delta, and Gamma variants. Top: WT footprints including the residues with an interaction within 4A of the partner. Middle: Visualization of the Delta variant interface. Bottom: Visualization of the Gamma variant interface. Mutations are labeled and represented as sticks. WT structures are superimposed for the RBD of each variant: WT, orange; variant, olive C. Cartoon illustrating additive and non-additive (epistatic) interactions between mutations. From left to right: additive, escape-neutral, escape-minimizing, and escape-exacerbating. Mi, Mj, Mij denote the effects of the single and double mutants.
Escape mutations less likely for Delta variant
The results of this study imply that the emergence of escape mutants is less likely for the SARS-CoV-2 Delta variant in comparison to the wild type strain and moderately more likely for the Gamma variant in comparison to the wild type strain as well.
More specifically, many mutations were found in six sites for the wild type SARS-CoV-2, arguably with the ability to substantially destabilize the complex of neutralizing antibodies and RBD relative to the RBD-ACE2 complex – posing, in turn, a risk of vaccine escape.
“Overall, little epistasis at the RBD interface was detected, with additive effects on the binding affinities observed for most pairs of mutations”, emphasize study authors in this bioRxiv paper.
Hence, the detected non-additive trends in the Delta variant weakly stabilize the interaction of the RBD with the neutralizing antibodies relative to the interaction with ACE2; conversely, epistasis is predicted to destabilize interaction more substantially with the neutralizing antibodies relative to ACE2 in the Gamma variant.
A view into the vaccine-escape landscape
The authors conclude that the small set of escape-enhancing mutations that have been already established for the wild-type SARS-CoV-2 strain likely include the majority of all possible mutations with this effect, which is a rather encouraging finding.
“The modest ensemble of mutations relative to the wild type that are currently known to reduce vaccine efficacy is likely to comprise the majority of all possible escape mutations for future variants,” say study authors.
Hence, this work opens the door for a cheap and rapid view into the vaccine-escape landscape for any emerging variant of concern; however, further studies are needed in order to increase the predictive power of such research endeavors.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources