A recent report published in the journal Cell Discovery has described the immunogenicity and protective efficacy of a novel coronavirus disease 2019 (COVID-19) subunit vaccine candidate. The vaccine induces robust immune responses in rodents and non-human primates and protects the animals against severe disease.
Background
The COVID-19 pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has imposed an unprecedented threat to humanity, with more than 224 million infections and 4.62 million deaths recorded worldwide. However, the fast-paced development of several potent vaccines has made it possible to tackle the pandemic to some extent. More than 300 vaccines, including live-attenuated, inactivated, viral vector-based, nucleic acid-based, and protein subunit vaccines, have been developed in record time and are currently under clinical investigations.
About 26 of these vaccines have already received emergency use approval by health authorities and are rolling out worldwide. However, the global mass vaccination campaigns are still struggling with the supply-demand mismatch despite this extraordinary achievement. This highlights the need for developing novel high-yield vaccine candidates with good safety and efficacy profiles.
In the current study, the scientists have developed a SARS-CoV-2 spike receptor-binding domain (RBD)-homodimer as a protein subunit vaccine and tested its immunogenicity and protective efficacy in rodents and non-human primates.
Development of RBD-homodimer
The RBD-homodimer was produced by covalently linking interdomain cysteine residues at position 538 via disulfide bonding. The homodimer was expressed in cells with a high yield and was subsequently purified for further characterization. The thermal stability analysis of the purified product revealed that the RBD-homodimer remained stable at 25 °C for at least 8 weeks. Moreover, the homodimer showed a high binding affinity for human angiotensin-converting enzyme 2 (ACE2), the host cell entry receptor of SARS-CoV-2.
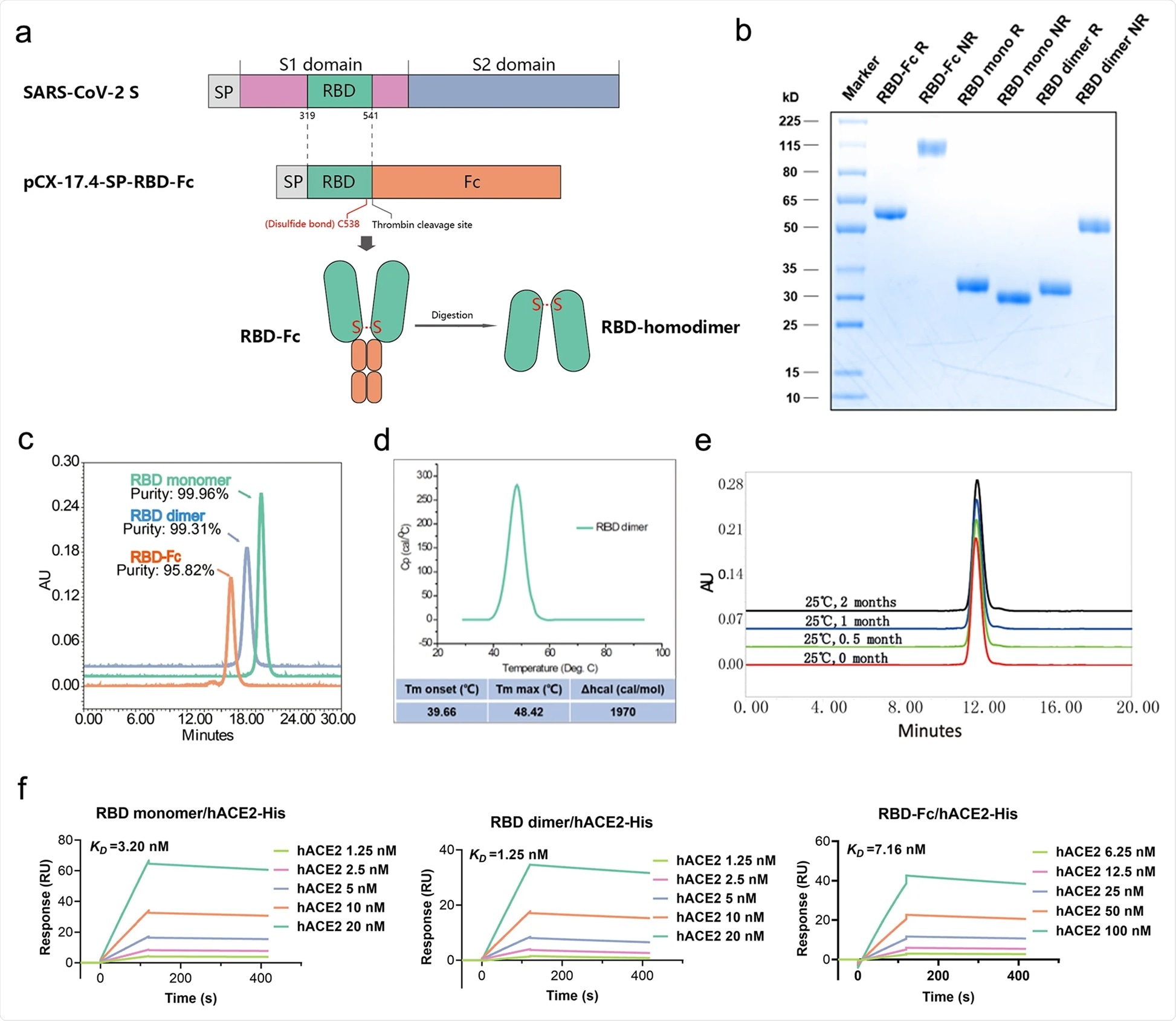
Schematic diagram of the full-length SARS-CoV-2 spike protein (S) and RBD protein (319–541 amino acid). Structural elements include cleavable signal sequence (SP, gray), S1 (pink), and S2 (blue), receptor binding domain (RBD, green), thrombin cleavage site, Fc (orange), disulfide bond at S protein position 538 amino acid between RBD monomers (red). b Analysis of the reduced and non-reduced RBD proteins and RBD protein precursors (RBD-Fc) through SDS-PAGE and Coomassie brilliant blue staining. RBD was acquired from thrombin digestion on RBD-Fc and removing Fc fragments. R reduced form, NR non-reduced form. c Analysis of RBD monomer, RBD dimer, and RBD-Fc proteins by size exclusion chromatography (SEC), and the corresponding purities were marked in the graph. d Thermostability of RBD dimer proteins calculated by differential scanning calorimetry (DSC). e The long-term stability of RBD dimer at 25 °C was detected by SEC. f The binding affinity of RBD monomer, homodimer, and RBD-Fc to hACE2-His. The corresponding KDs were calculated using the Biacore T200 Evaluation 3.0 (software) with “1:1 binding” model as the curve fitting method.
Protective efficacy of RBD-homodimer in mice
For immunogenicity profiling, mice were immunized with three doses of RBD-homodimer at 2-week intervals. The vaccine exhibited the highest efficacy in inducing robust anti-RBD binding and neutralizing antibodies after the third immunization. Regarding cellular immune response, the vaccine showed high ability in inducing RBD-specific T cells secreting IL-4, IFN-γ, and IL-2.
In a separate set of experiments, human ACE2-expressing mice were vaccinated first and subsequently challenged with SARS-CoV-2 intranasally. The in vivo analysis revealed that the vaccine gradually reduces the viral loads to an undetectable limit by day 5 post-infection. Similarly, no infectious virus was detected in vaccinated mice at day 5 post-infection. In addition, the histopathological analysis revealed that the vaccinated mice have only mild pulmonary pathological signs with almost no lung infiltration of inflammatory cells at days 2 and 5 post-infection. In contrast, unvaccinated mice showed moderate to severe lung pathologies after infection.
Protective efficacy of RBD-homodimer in non-human primates
Rhesus macaques were immunized with three doses of the vaccine intramuscularly, followed by viral challenge at day 42 post-first immunization. All vaccinated monkeys showed robust antibody responses similar to vaccinated mice. The cellular immune response to the vaccine in monkeys was also comparable to that in mice.
At day 7 post-infection, none of the vaccinated monkeys showed detectable levels of viral RNA in respiratory swabs. Similarly, no viral RNA was detected in anal swabs. Compared to unvaccinated monkeys, vaccinated monkeys showed significantly less severe pulmonary lesions.
Cross-reactive immunity
The cross-reactivity of RBD homodimer-induced antibodies was tested against nine RBD variants with mutations at K417N, N439K, Y453F, S477N, T478I, G485S, F490S, S494P, and N501Y. The antibodies showed a binding affinity for all tested mutants, with slightly reduced reactivity against the K417N, N439K, and T478I mutants. Moreover, the ACE2 – RBD blocking assay findings revealed that vaccine-induced antibodies are capable of inhibiting the ACE2 – RBD interactions and neutralizing tested RBD mutants. However, the highest and lowest levels of antibody-mediated neutralization were observed against the N501Y and K417N mutants, respectively.
The neutralization assay conducted against the wildtype SARS-CoV-2 and the beta variant (first originated in South Africa) revealed that vaccine-induced antibodies have 1-fold lower neutralizing efficacy against the beta variant compared to that against the wildtype virus.
Study significance
Collectively, the study reveals that the novel RBD-homodimer vaccine candidate has good immunogenicity and protective efficacy against severe COVID-19. Importantly, the vaccine can induce cross-reactive antibodies against more deadly SARS-CoV-2 variants.