As of September 12, 2021, there have been more than 4.62 million deaths due to the coronavirus disease 2019 (COVID-19), which is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). SARS-CoV-2 is a highly communicable virus that causes pathology of the respiratory system shortly after its entry into host cells. Positioned on the surface of SARS-CoV-2 is the spike (S) glycoprotein, which must bind to the human host receptor angiotensin-converting enzyme 2 (ACE2), to initiate viral entry into host cells.
 Study: Network analysis outlines the strengths and weaknesses of emerging Spike variants. Image Credit: ktsdesign / Shutterstock.com
Study: Network analysis outlines the strengths and weaknesses of emerging Spike variants. Image Credit: ktsdesign / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Structure of SARS-CoV-2
The SARS-CoV-2 S protein is comprised of three identical monomers (homotrimer) and is a class I viral fusion protein. Moreover, the S protein has the ability to appropriate various conformations that are dependent on the formation of the receptor-binding domain (RBD) of its protomers.
There are two dominant confirmations that are adopted by the S protein. These include the all down confirmation, in which all protomers are in a closed state, or the one up confirmation, in which one protomer is open and two are closed.
When compared to other viruses that have spread across the world, SARS-CoV-2 has demonstrated a slow rate in the accumulation of immunological significant variants, which has been seen in other coronaviruses. While this may be true, the SARS-CoV-2 S protein has been the location for the most notable evolutionary changes. A single amino acid substitution in residue 614 of the S protein, for example, resulted in the G form variant, which replaced the original D form variant as the dominant strain.
The D614G variant resilience to communication disruption
The variation displayed in the communication core through the networks emphasizes the influence of the D614G variant shift in the one up and all down states. At the base of the RBD, the ring straddles the central plate of the spike protein at the starting point of the N-terminal domain (NTD) and extends via the C-terminal domain 0 (CT0).
The high centrality band of residues on the S protein extend at the lowest point of the furin cleavage hairpin to the beta-strands and incorporates residues of the CT0, C-terminal domain 1 (CT1), C-terminal domain 2 (CT2), and NTD, which occurs in both the D and G variants. However, in comparison with the D form, the G form communication ring is compromised with a lower number of CT2 residues and a higher number of CT0 residues. Despite these structural changes, this variant exhibits increased stability with the fraction of residues situated at each region.
For communication between the outer domains of the S protein, the core ring is vital. The NTD supersite, which is avoided by the high centrality core, is the site of where many antibodies have displayed affinity and effectiveness.
The influence of each of the aforementioned domains incorporated in the core ring have been shown to play key roles in the functionality of the S protein. Allosteric linkage is displayed by the moving components of the S protein, thereby demonstrating that protein function is not hindered by a site of preferential antibody binding.
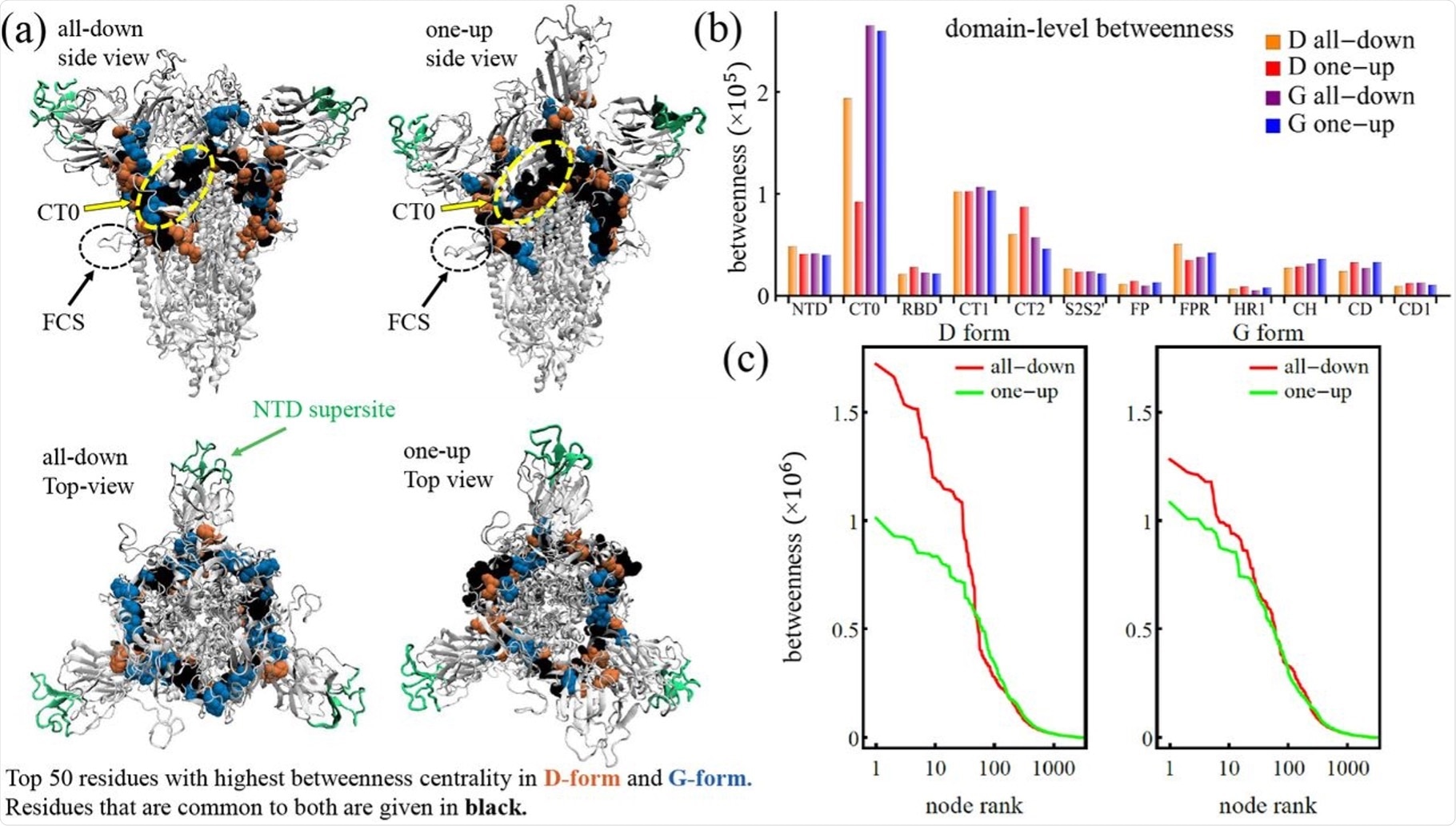 The communication core of the spike protein. (a) Spike protein with NTD supersite highlighted and top 50 residues of the D and G forms. (b) Domain level betweenness broken down to D and G form. (c) Individual residue betweenness centrality.
The communication core of the spike protein. (a) Spike protein with NTD supersite highlighted and top 50 residues of the D and G forms. (b) Domain level betweenness broken down to D and G form. (c) Individual residue betweenness centrality.
Creating a more balanced in the G-form variant
The RBD is a spherical domain that switches between open and closed conformations. Notably, the ACE2 binding site is only considerably exposed when the RBD is in the open conformation. Functional adjustment of two loops at the C-terminal and N-terminal cause the RBD transition between open and closed to occur, with the loops operating as hinges to allow the RBD to swivel.
The G-form in the open conformation achieves greater stability when utilizing the hinge residues as compared to the D-form. This increased stability in the G-form allows for a greater balanced allosteric communication pathway by using both hinges, thus giving a more robust response to any mutations that may arise.
Critical communication ring of S protein
Within the S protein, a vital communication ring has been identified which connects the peripheral regions, such as the furin cleavage sites, the NTD supersite, and the open RBD, together with but also to the core of the S protein core. Notably, the core of the S protein is the site in which vital processes occur during membrane fusion.
The CT0, CT1, CT2, and NTD domains are the main structural components of the communication ring. These regions of the communication ring also display the capacity to impact the entire protein network, which consists of the pathways originating from these regions that can reach the entire area of the protein more efficiently. The NTD, which is the dominant region of the communication ring, is situated next to and in contact with the open RBD, which suggests that the NTD functions to facilitate the binding of RBD or the initial attachment of the virus to the host cell.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Manrique, P. D., Chakraborty, K., Nguyen, R., et al. (2021). Network analysis outlines the strengths and weaknesses of emerging Spike variants. bioRxiv. doi:10.1101/2021.09.03.458946. https://www.biorxiv.org/content/10.1101/2021.09.03.458946v1.full.
- Peer reviewed and published scientific report.
Manrique, Pedro, et al. "Network Analysis Uncovers the Communication Structure of SARS-CoV-2 Spike Protein Identifying Sites for Immunogen Design." IScience, vol. 26, no. 1, 2023, p. 105855, https://doi.org/10.1016/j.isci.2022.105855, https://www.sciencedirect.com/science/article/pii/S2589004222021289