The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) triggered the ongoing coronavirus disease 2019 (COVID-19) pandemic that brought life to a grinding halt in much of the inhabited world for most of 2020. As this pandemic continues to cause thousands of new infections and deaths worldwide each day, researchers are earnestly seeking to understand how SARS-CoV-2 works to infect host cells and inactivate host defenses.
Background
SARS-CoV-2 is a large RNA virus that causes both human and animal infection. Although SARS-CoV-2 generally is associated with its respiratory manifestations, in severe and critical cases, it induces overwhelming inflammation that results in multi-organ dysfunction and even death.
The current study involves microRNAs (miRNAs), which are about 22 nucleotides in length. These miRNAs comprise non-coding RNA molecules (ncRNAs) that modulate gene expression in the post-transcriptional stage. This action is mediated by partial inhibition or mRNA decay.
The formation of miRNAs
While messenger RNA (mRNA) is being synthesized in the cytoplasm, the enzyme RNA polymerase II causes primary miRNA molecules to be transcribed, all of which have a characteristic tendency to form strong hairpins. The endonuclease Drosha recognizes such hairpin sequences and cleaves them, leading to the production of miRNA precursors that are about 70 nucleotides in length.
These precursor molecules are then trafficked into the cytoplasm where they are cleaved by another enzyme known as Dicer, which assists in the formation of duplex miRNAs that are 22 nucleotides in length. These become the cargo of host Argonaute (Ago) proteins.
At this point, the passenger strand is removed. The Ago proteins bind to target sequences that are partly complementary to the miRNA. These sequences are mostly in the 3′ untranslated region (UTR) of mRNAs. This binding causes inhibition of translation of these mRNAs or their decay.
When the target mRNA and binding miRNA sequences are almost completely complementary, the Ago2 protein cleaves the mRNA. This cleavage is an action that is more characteristic of small interfering RNA molecules (siRNAs).
Both these functions are part of the RNA interference (RNAi) pathway. However, Ago2-mediated RNA cleavage occurs at a lower copy number and silences the target gene more powerfully than the standard miRNA mode of action.
It is interesting that the number and types of miRNAs differ by the host cell and stage of development. Also, each miRNA is capable of reducing the level of expression of a range of transcripts. The outcome is a precise tuning of gene expression of almost all mRNAs as required by each type of cell at its particular stage.
Viruses and miRNAs
The presence of abnormal miRNAs is typically a marker of disease. Very often, this is the case with a viral infection, as viruses take over this mechanism to reduce the level of host miRNAs or produce their own miRNAs.
Herpes viruses, for instance, transcribes certain sequences that lead to the selective decay of some host miRNAs through a process known as target-directed miRNA degradation (TDMD). The poxvirus poly(A) polymerase causes extensive miRNA polyadenylation that ultimately leads to miRNA decay.
Viruses can also use unconventional non-RNAi pathways to avoid the need for Drosha, rather than synthesizing their own miRNAs in the cytoplasm. Coronaviruses such as SARS-CoV-2 may be of this kind, producing small viral RNAs (svRNAs) as part of their disease-producing activity, including miRNA-like strands that trigger inflammation and type I interferon (IFN) signaling.
SARS-CoV-2 and vmiR-5p
The current study reports the discovery of a viral miRNA called vmiR-5p, which is a miRNA-like viral ncRNA that is expressed by SARS-CoV-2.
SARS-CoV-2 does not appear to affect the host miRNAs to an appreciable extent, as shown by the tiny number of small RNA sequences obtained from infected host cells. A potentially valuable observation is that after SARS-CoV-2 infection, the miRNA abundance was increased, whereas two other small ncRNA classes were reduced.
In other words, the virally-induced shut-off of host transcripts does not affect host miRNA production, perhaps because the latter are protected within circulating exosomes. This points to the potential for using small RNAs to treat COVID-19.
Among the viral miRNAs, about 5% were 20-nucleotide sequences related to the ORF7a, which is a viral gene that encodes one of the viral accessory proteins and is implicated in innate immunity. The protein is thought to be an antagonist of the type I IFN response.
Deletions of ORF7a are associated with decreased innate immune evasion. However, these deletions chiefly affect the C-terminal end of the protein, while leaving the N-terminal end, which contains vmiR-5p, intact.
This ncRNA is part of a strong hairpin that is highly conserved in coronaviruses. The presence of this hairpin identifies it as a miRNA since hairpin formation occurs during the biogenesis of miRNAs.
This vmiR-5p sequence is found in SARS-CoV-2 infection and is positively related to the viral load/genomic RNA. Secondly, the researchers found that vmiR-5p binds to Ago proteins and may silence host target transcripts. Thus, even though vmiR-5p is expressed at low levels, its association with Ago proteins exploits sequence complementarity that can cause significant target mRNA cleavage while tolerating a small number of mismatched nucleotides.
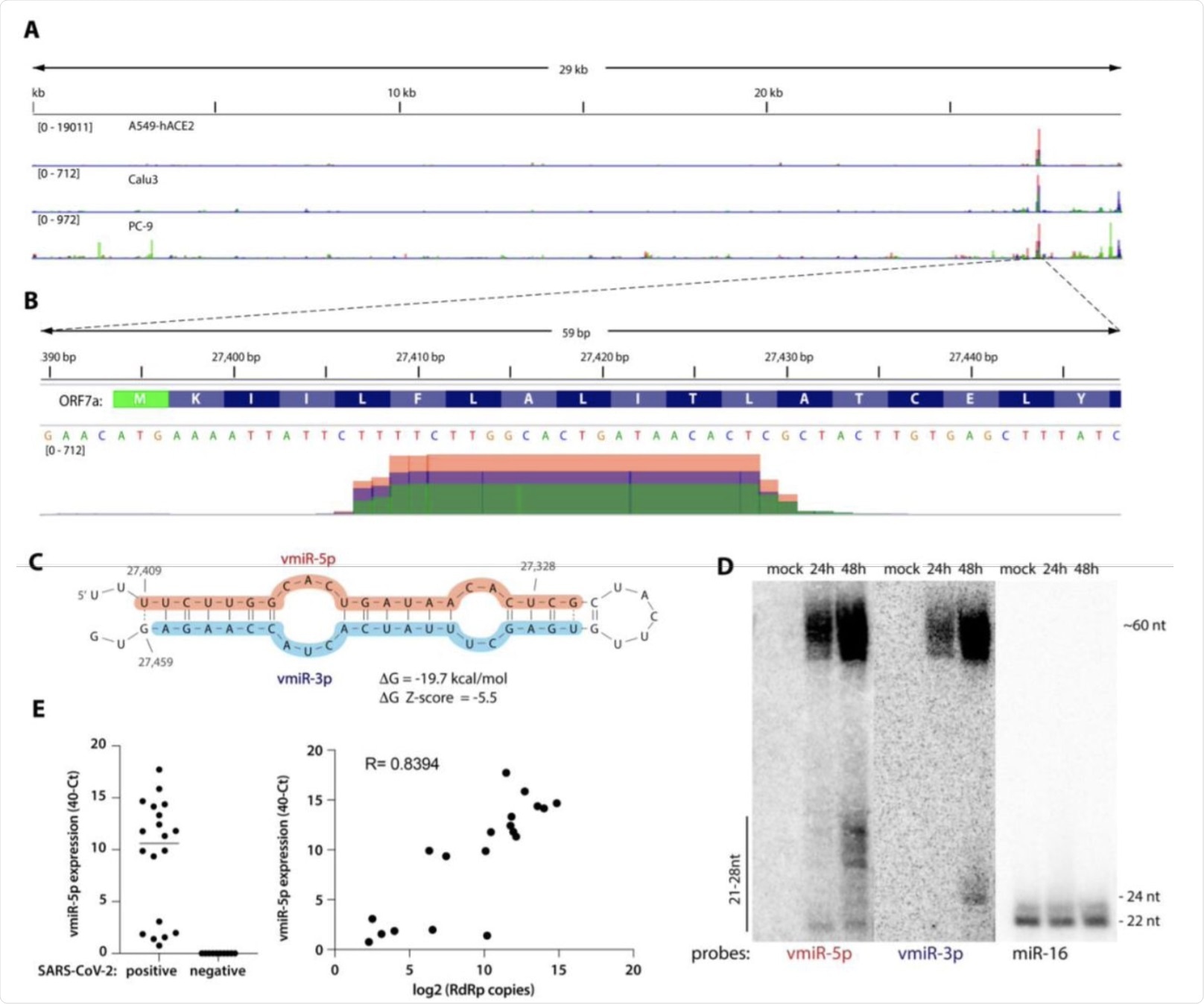 A) Viral small RNA reads obtained from the three cell lines infected with SARS-CoV-2 map to a single distinct peak within the viral genome (data for MOI 5, 24 hpi are shown). The replicates for each cell line were overlayed on a single track (represented by different colors) and are normalized to 107 total reads. MOI — multiplicity of infection. B) The reads coming from SARS-CoV-2 (∼20 nt-long) map near the beginning of the ORF7a gene (encoded amino acids are shown above the nucleotide sequence). Data from Calu-3 cells are shown. C) vmiR-5p forms a hairpin with the sequence immediately downstream in the viral genome. Shaded nucleotides indicate the sequences detected by Northern blot probes: pink for 5p and blue for 3p. D) vmiR-5p can be detected by Northern blotting of extracts from Calu-3 cells infected with SARS-CoV-2 at MOI 0.05. E) As measured by custom TaqMan RT-qPCR, vmiR-5p is present in nasopharyngeal samples from SARS-CoV-2-infected individuals (right panel), with its levels correlating with viral load (left panel). RdRp — RNA-dependent RNA polymerase.
A) Viral small RNA reads obtained from the three cell lines infected with SARS-CoV-2 map to a single distinct peak within the viral genome (data for MOI 5, 24 hpi are shown). The replicates for each cell line were overlayed on a single track (represented by different colors) and are normalized to 107 total reads. MOI — multiplicity of infection. B) The reads coming from SARS-CoV-2 (∼20 nt-long) map near the beginning of the ORF7a gene (encoded amino acids are shown above the nucleotide sequence). Data from Calu-3 cells are shown. C) vmiR-5p forms a hairpin with the sequence immediately downstream in the viral genome. Shaded nucleotides indicate the sequences detected by Northern blot probes: pink for 5p and blue for 3p. D) vmiR-5p can be detected by Northern blotting of extracts from Calu-3 cells infected with SARS-CoV-2 at MOI 0.05. E) As measured by custom TaqMan RT-qPCR, vmiR-5p is present in nasopharyngeal samples from SARS-CoV-2-infected individuals (right panel), with its levels correlating with viral load (left panel). RdRp — RNA-dependent RNA polymerase.
Putative functions
The scientists of the current study could not quantitatively assay the effect of vmiR-5p on mRNA transcription within infected cells due to the large-scale suppression of host transcription. Using synthetic vmiR-5p, they found that two predicted target host mRNAs were downregulated.
One of these downregulated mRNAs is the Basic Leucine 18 Zipper ATF-Like Transcription Factor 2 (BATF2), which is involved in IFN-gamma signaling. The second mRNA was Heparan Sulfate Proteoglycan 2) HSPG2, which may be implicated in viral superinfection.
Other possible functions of the vmiR-5p may include the regulation of viral subgenomic or antigenomic RNA. Additional functions of the hairpin itself remain to be elucidated.
Though this miRNA is produced through the host cell’s machinery, it occurs by the processing of a strong hairpin created within the viral ORF7a sequence. This processing requires neither other viral proteins nor Drosha, but may be enhanced by viral genes.
Conclusion
“Viruses develop multiple ways to suppress host gene expression, 20 and multiple overlapping mechanisms often evolve.”
The current study reveals a new strategy used by SARS-CoV-2, in addition to its known destabilization of host mRNA, its inhibition of translation, splicing, and export. This RNAi-dependent pathway selectively silences host transcripts, and perhaps even viral transcripts, thus regulating gene expression as necessary for optimal viral replication.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Pawlica, P., Yario, T. A., White, S., et al. (2021). SARS-CoV-2 Expresses a MicroRNA-Like Small RNA Able to Selectively Repress Host Genes. bioRxiv. doi:10.1101/2021.09.08.459464. https://www.biorxiv.org/content/10.1101/2021.09.08.459464v1
- Peer reviewed and published scientific report.
Pawlica, Paulina, Therese A. Yario, Sylvia White, Jianhui Wang, Walter N. Moss, Pei Hui, Joseph M. Vinetz, and Joan A. Steitz. 2021. “SARS-CoV-2 Expresses a MicroRNA-like Small RNA Able to Selectively Repress Host Genes.” Proceedings of the National Academy of Sciences 118 (52): e2116668118. https://doi.org/10.1073/pnas.2116668118. https://www.pnas.org/doi/full/10.1073/pnas.2116668118.