Coronaviruses such as the severe acute respiratory syndrome coronavirus (SARS-CoV) and SARS-CoV-2 are enveloped positive-sense RNA viruses that are a huge threat to public health globally. They can be transmitted from animals to humans and can cause many diseases ranging from the common cold to severe respiratory illnesses.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Structural similarities between SARS-CoV and SARS-CoV-2 viral proteins
The SARS-CoV and SARS-CoV-2 viruses have four main structural proteins: the spike (S), nucleocapsid (N), envelope (E), and membrane (M) proteins. Cryo-electron microscopy (cryo-EM) structures of SARS-CoV-2 spike protein show that it shares architecture with SARS-CoV spike protein, and the sequence of SARS-CoV-2 S is about 77% same as that of SARS-CoV S. Both spike proteins recognize and bind to angiotensin-converting enzyme 2 (ACE2) in the host cells in order to enter the host and infect them.
Despite the similarities, the toxicity and transmissibility of SARS-CoV and SARS-CoV-2 are significantly different. Given the global health emergency caused by the COVID-19 pandemic, there is a pressing need to clarify the function of the envelopes of the coronaviruses and analyze why they have different transmission capabilities.
Combining cryo-EM and structure prediction data to build coarse-grained models of SARS-CoV and SARS-CoV-2 envelopes
Newer structures of SARS-CoV-2 proteins are being obtained at regular intervals using cryo-EM. Computational approaches are also being used for structural prediction of molecular dockings of various molecules on viral proteins and free energy calculations of the spike-ACE2 binding process. However, larger-scale spatio-temporal processes, including virus architecture, fusion, and budding, and virion assembly, are still not fully understood and are a challenge for experimental techniques and molecular dynamics simulations.
Researchers from China and Canada recently combined structural data obtained from cryo-EM and structure prediction and constructed bottom-up Martini coarse-grained models of intact envelopes of SARS-CoV and SARS-CoV-2. They performed microsecond molecular dynamics simulations that explored the dynamics and supramolecular organization of the viral envelopes. This study is available on the bioRxiv* preprint server.
Results show marked differences in flexibility and interaction patterns of spike proteins of SARS-CoV and SARS-CoV-2 envelopes
The results showed that both SARS-CoV and SARS-CoV-2 envelopes have a spherical morphology, with the structural proteins forming multiple string-like islands in the membrane and clusters between spike protein heads.
“Previous experiments have confirmed that nanocluster is required for efficient pathogen binding and internalization.”
The main differences between the envelopes of SARS-CoV and SARS-CoV-2 are the flexibility of the spike proteins and the interaction patterns between the spike proteins. The researchers believe that the models used in this study offer structural and dynamic insights into the SARS coronavirus envelopes and could help further investigations.
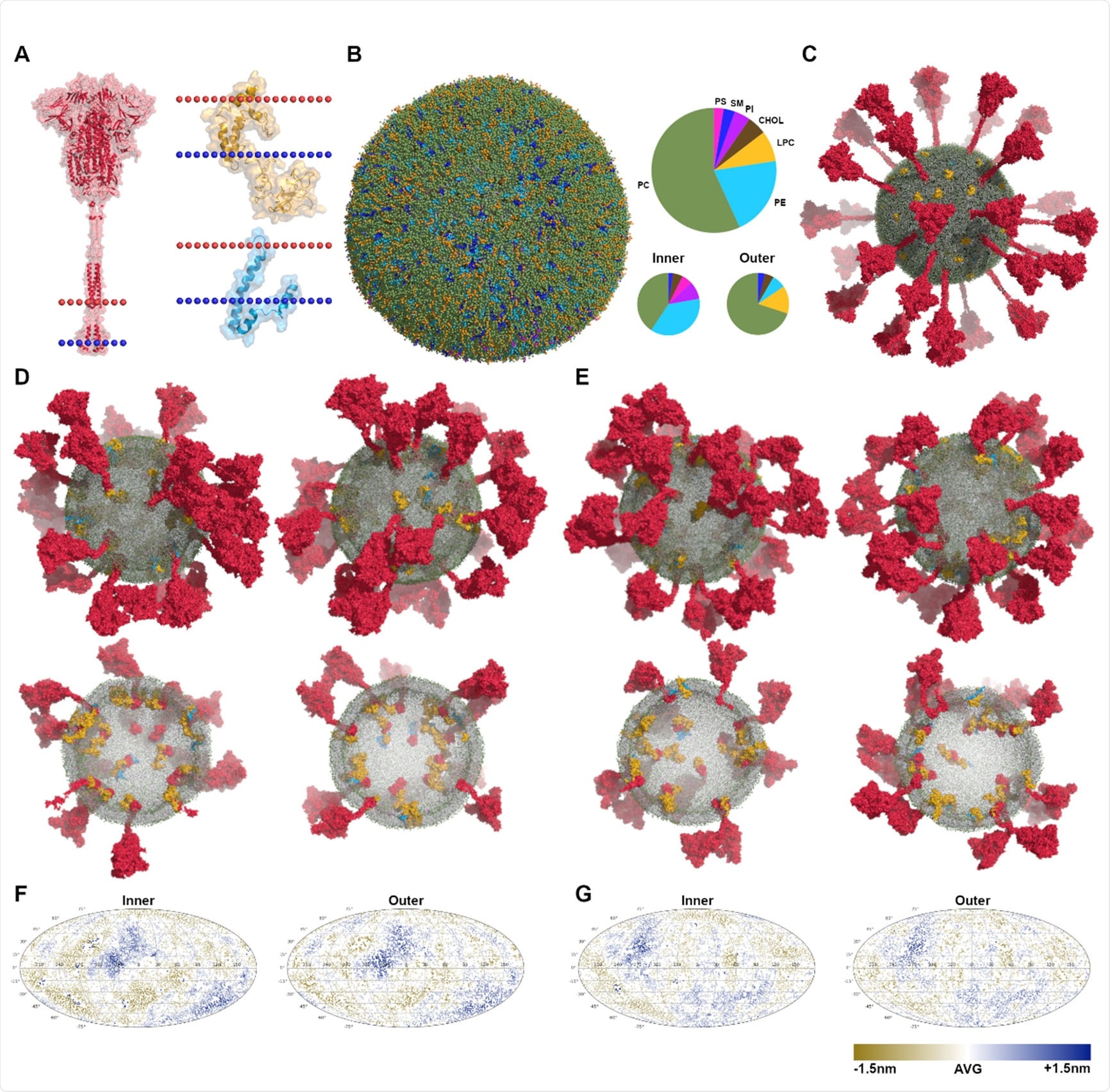
Martini CG models of virus envelopes. (A) The structures of S (in red), M (in yellow) and E (in blue) with the dotted lines indicating the position of lipid bilayers. (B) The equilibrated vesicle with the lipid composition according to that of the endoplasmic reticulum (ER). The lipid bilayer is asymmetric, the compositions of inner and outer leaflets are different. Lipid types considered here are phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine (PS), lyso-PC (LPC), sphingomyelin (SM), phosphatidylinositol (PI), and cholesterol (Chol). (C) Multiple copies of proteins (E×15 in blue, S×30 in red, and M×45 in yellow) were inserted into the equilibrated vesicle, making up the initial CG model of virus envelopes. (D)-(E) The equilibrated models after microsecond MD simulations of SARS-CoV-2-md1 and SARS-CoV-md3, illustrated by the side view and transverse section. (F)-(G) Mollweide projection maps of the envelope membranes of SARS-CoV-2-md1 and SARS-CoV-md3, illustrated using the distances between lipid heads and the vesicle’s center of mass.
Structural findings offered by these models facilitate better understanding of the viral envelopes
According to the authors, their Martini CG models illustrate the experimental complexity and scale of SARS-CoV and SARS-CoV-2 envelopes at the atomistic level. The structural proteins of these viruses are not uniformly distributed across the viral envelopes. Most spike proteins form oligomers with extensive interactions between their heads, while the transmembrane domains of the proteins cluster into string-like islands, mediated by lipids that are negatively charged.
“The interaction patterns and flexibility of S stalks show agreements with the experimental observations and all-atom simulations, indicating the reliability of the structural and dynamic details obtained from our simulations.”
The simulations used in this study revealed that the critical difference between SARS-CoV and SARS-CoV-2 envelopes is in the S-S interaction patterns and the intrinsic flexibilities of the spike proteins. SARS-CoV-2 Ss are more inclined to interact with each other and have higher inherent flexibility to recognize and bind to the ACE2 receptors compared to SARS-CoV spike proteins. These may be two of the key reasons why SARS-CoV-2 is more infectious than SARS-CoV. The structural and dynamic details provided by these models offer a better understanding of the viral envelopes and can help drug development in the future.
“In the simulations, we are able to observe the forming of S clusters and supramolecular islands by the transmembrane domains of different proteins and lipids in the envelope membrane.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources