A team of scientists from the USA and France has recently unveiled the mechanism of immune dysregulation in coronavirus disease 2019 (COVID-19) patients by analyzing the early-stage interaction between severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-infected epithelial cells and immune cells. The study is currently available on the bioRxiv* preprint server.
Aberrant activation of the inflammatory network is one of the major hallmarks of severe COVID-19, which can subsequently result in overwhelming production of cytokines, disrupted interferon signaling, alterations in myeloid cell and T cell functions, and production of autoantibodies. Suppression of interferon signaling at the initial phase of infection can potentiate viral replication, leading to the augmentation of immunopathological signs.
Regarding SARS-CoV-2 pathogenicity, it is generally considered that the viral genome is equipped to disrupt and escape host immune responses, leading to severe immunopathology.
In the current study, the scientists have aimed to identify the factors responsible for impaired immune responses COVID-19.
They developed an in vitro co-culture system to study the interaction between immunocytes and SARS-CoV-2-infected epithelial cells at the early infection stage. Futher, they characterized the co-cultured immunocytes by transcriptomics and flow cytometry.
Important observations
For profiling, the scientists selected monocytes and B cells as they are the first-line sensors of infection and are known to have dysregulated transcriptomes in COVID-19 patients. They observed a strong monocyte response characterized by 675 differentially expressed genes. Specifically, monocyte responses were characterized by induction of pro-inflammatory cytokine and chemokine genes and interferon-stimulated genes and downregulation of class II major histocompatibility complex (MHC) genes.
With further analysis, they observed that exposure to virus-infected epithelial cells induces significant alterations in monocytes in terms of cytokine production, innate immune signaling, cell mobility and adhesion, and antigen presentation. In contrast, they observed only a mild transcriptomic effect of SARS-CoV-2 directly on infected epithelial cells.
By analyzing B cell populations, they observed a partially similar response as monocytes; however, the magnitude of antiviral response was highest in monocytes.
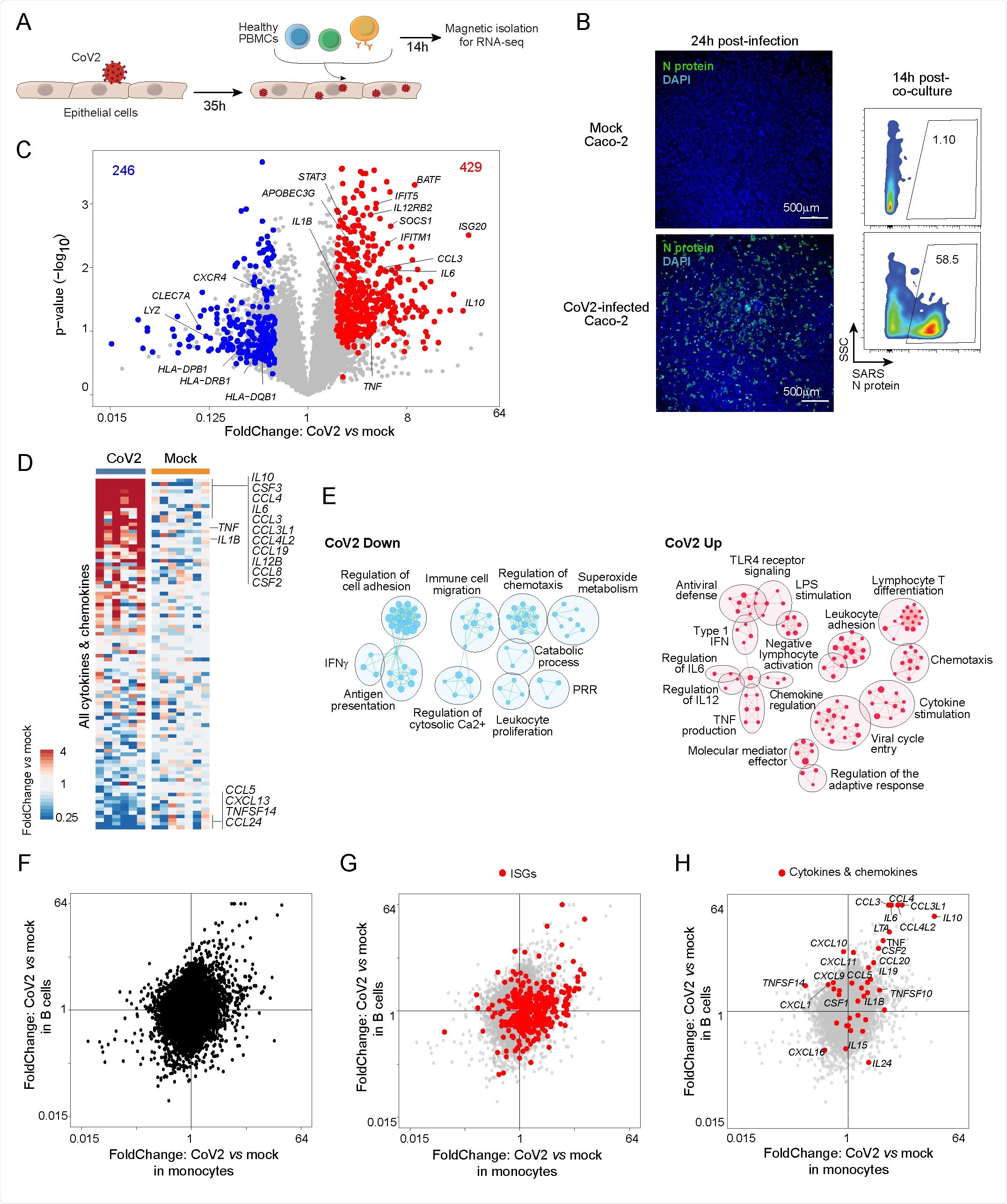
SARS-CoV2 epithelial-immune co-culture triggers a mixed inflammatory response in immunocytes. (A) Experimental approach. Caco-2 monolayer cultures were infected with CoV2 for two hours. Thirty-five hours post-infection, the monolayer was washed twice and PBMCs from HDs were added directly to the cultures. These were harvested 14 h later, and subpopulations (CD14+ monocytes and CD19+ B cells) were magnetically purified for population RNAseq. (B) Infection rate in Caco-2 cells assessed by immunofluorescence with an anti-SARS-CoV2 N antibody: displayed by microscopy at 1 day post-infection (upper panel, 4x, N antibody + antirabbit-AF488 + DAPI) and by flow cytometry at the time of harvesting PBMCs (~48 h postinfection, lower panel). (C) FoldChange vs p-value (volcano) plot of gene expression in monocytes co-cultured with CoV2 infected Caco-2 compared to monocytes co-cultured with uninfected Caco-2 (mock). Genes from CoV2 up signature (red) and CoV2 down signature (blue) are highlighted. (D) Heatmap of the expression of cytokine transcripts in CoV2 versus Mock condition (as ratio to mean of mock for each condition and each experiment). (E) Gene ontology analyses of CoV2 up (right, red) and down (left, blue) signatures displayed as an enrichment map. Pathways are shown as circles (nodes) that are connected with lines (edges) if the pathways share many genes. Size of the node is proportional to the number of genes included in this pathway. (F-H) FoldChange-FoldChange plots comparing the response in monocytes (x-axis) relative to B cells (y-axis) in the context of CoV2 Caco-2 infection, without highlight (F) or highlighted with: interferon-stimulated genes (ISGs) (G); cytokine and chemokine transcripts (H).
Inflammatory responses induced by SARS-CoV-2, influenza virus, and Ebola virus
In this section, the scientists exposed monocytes to epithelial cells infected with either of the three viruses. By analyzing transcriptional changes in monocytes, they observed that both SARS-CoV-2 and influenza virus share a similar profile of downregulated genes. However, for upregulated genes, they observed both shared and virus-specific alterations. Given the significant downregulation of eosinophil chemotactic factor CCL24 observed in the analysis, they suggested that eosinophil recruitment is highly disrupted in both SARS-CoV-2 and influenza infections.
The comparison between SARS-CoV-2 and Ebola virus infections revealed a similar trend, with some shared effects and some virus-specific effects. Similar to that observed in influenza virus comparison, the induction of inflammatory cytokines and chemokines TNF, IL1B, CCL3 and IL10 was found to be highly specific to SARS-CoV-2 infection.
Given these observations, the scientists suggest that some changes observed in monocytes are generic to all tested infections. However, some responses, especially pro-inflammatory changes, are highly specific to SARS-CoV-2 infection.
SARS-CoV-2 proteins involved in monocyte induction
The scientists studied the impact of a panel of viral proteins and identified two distinct sets of upregulated and downregulated genes. Specifically, they observed that the spike protein and non-structural proteins (NSP) 5, 9, and 14 strongly upregulate pro-inflammatory genes in co-cultured monocytes.
With further analysis, they found that the changes in monocytes are not directly caused by the viral proteins. Instead, the changes induced in infected epithelial cells by the viral proteins are responsible for altering monocyte response.
Importantly, they observed only a mild transcriptional effect of SARS-CoV-2 proteins in infected epithelial cells. With further analysis, they concluded that SARS-CoV-2-induced epithelial responses are mediated by soluble factors.
Clinical relevance of the study
The scientists analyzed the gene expression data of myeloid cells isolated from severe COVID-19 patients to determine whether observed in vitro changes can be correlated with the in vivo changes observed in COVID-19 patients.
Their analysis revealed that the SARS-CoV-2 signature observed in the co-culture model is similar to the dysregulated myeloid response observed in patients both at local and systemic levels.
Importantly, they observed that monocytes isolated from children are not responsive to SARS-CoV-2. Specifically, the co-culture of monocytes from children with virus-infected epithelial cells did not cause any significant changes in the expressions of cytokine/chemokine genes and interferon-stimulated genes in monocytes. These findings justify the relatively lower susceptibility of children to severe COVID-19.
Study significance
The study indicates that interactions between epithelial and immune cells at the earliest stage of infection might be responsible for lethal inflammatory responses in severe COVID-19 patients. As mentioned by the scientists, therapeutic modulation of these early interactions could reduce COVID-19 severity.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources