Researchers in the Republic of Korea have conducted a study of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – the agent that causes coronavirus disease 2019 (COVID-19) – that could help to inform further studies investigating the increased virulence of variants of concern and interest.
The team – from the Korean Research Institute of Chemical Technology in Daejeon – demonstrated that the four currently circulating variants of concern exhibit differences in plaque size, thermal stability and replication rates.
Thermal stability correlated with plaque size in all variants except the B.1.1.7 (alpha) lineage.
The largest mean plaque size was observed for the B.1.351 (beta) variant, followed by P.1 (gamma), B.1.617.2 (delta), and alpha.
Young-Chan Kwon and colleagues suggest that the relatively large plaque size observed for beta may account for its increased thermal stability, which may, in turn, contribute to its increased transmissibility.
The researchers say these observations may help characterize the variants in terms of their interactions with host factors and responses to environmental conditions.
Such studies will be crucial to informing effective management of the COVID-19 pandemic, they add.
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
More about the currently circulating variants of concern
Since the COVID-19 pandemic began in late December 2019, the genetic evolution of SARS-CoV-2 has resulted in the emergence of several novel variants that are more virulent and infectious than the ancestral strain. Four of these variants (alpha, beta, gamma, and delta) have been classified as variants of concern by the World Health Organization.
These four variants share the D614G mutation that confers increased infectivity, probably due to changes that affect the virus’s ability to bind to and fuse with host cells. The N501Y mutation that is shared by alpha, beta, and gamma is also known to increase binding affinity for host receptors and, therefore, cellular entry.
However, the particular combination of mutations that a variant has acquired could result in more significant conformational changes and distinctive modifications, says Kwon and colleagues.
For example, variants of concern exhibit differential receptor-binding affinity, with alpha requiring the most force to detach it from host cell receptors, followed by beta, gamma and delta.
What did the researchers do?
In the current study, the researchers found that each of the four variants of concern exhibited differences in plaque size, with the largest mean size observed for beta, followed by gamma, delta, and alpha.
While there are numerous determinants of plaque size, the researchers hypothesized that differences in the receptor-binding affinity, thermal stability, and replication rate of these viruses were likely contributors.
The team assessed the thermal stability of the variants by measuring their infectivity and half-life following prolonged incubation (2, 4, 12 and 24 hours) at different temperatures. Infectivity was measured using a focus-forming assay.
Of the four variants, beta exhibited the highest thermal stability, with a half-life approximately twice that of gamma or delta.
Correlation analyses revealed a strong association between plaque size and thermal stability in all variants except for alpha. This variant had a relatively long half-life, despite having a small plaque size.
The researchers say the results suggest that the relatively large plaque size observed for the beta variant may account for its increased thermal resistance.
“This increased stability may contribute to its pathobiology and transmission, requiring further studies of the viral titer and case fatality rate in humans,” they write.
Examining viral replication rates of the variants
Next, the team examined the viral replication rates of the four variants in Vero E6 cells. A plaque-forming assay was used to assess infectious viral particle numbers and quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was used to assess viral RNA concentration.
The alpha variant had fewer infectious viral particles than all the other variants and alpha and gamma had the lowest concentration of viral RNA.
The results suggest that although the alpha variant exhibits a relatively high thermal stability, its viral replication rate is probably low, thereby contributing to its small plaque size.
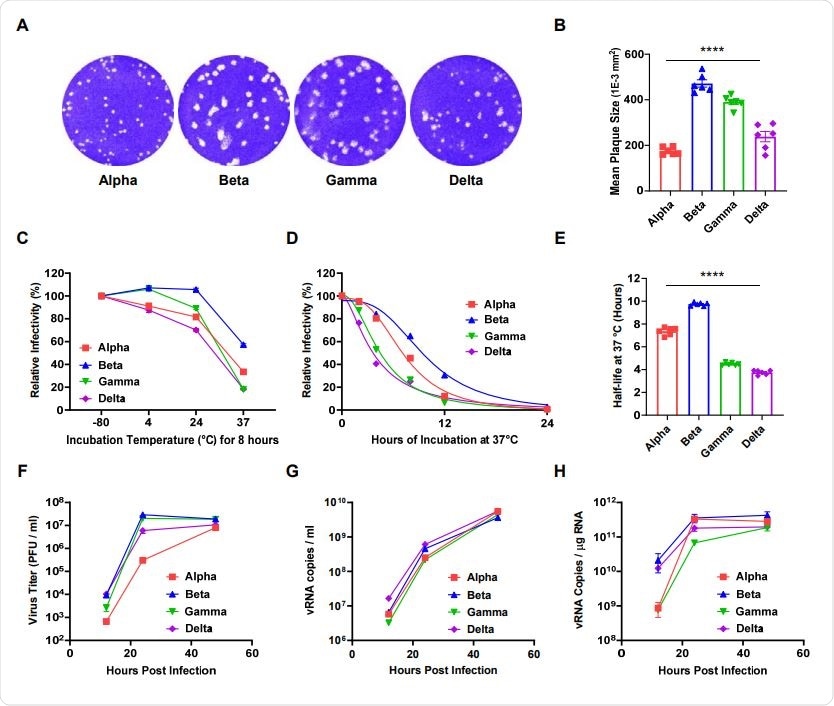
Comparative analyses of the plaque size, thermal stability, and replication rate of four SARSCoV-2 variants. (A) Representative images of the plaque-forming assay for each variant of concern. (B) The mean plaque size (1E-3 mm2) of each variant. (C) The relative infectivity of each variant after incubation at 4, 24, or 37 °C for 8 h as evaluated via focus forming assay. (D) Nonlinear regression of the relative infectivity of each variant following prolonged incubation (2, 4, 8, 12, and 24 h) at physiological temperatures (37 °C). (E) Average half-life values for each variant at 37 °C. (F) The infectivity of the progeny viruses was evaluated via plaque-forming assay completed at 12, 24, and 48 h post-infection. (G) Extracellular and (H) intracellular viral RNA was assessed by qRT-PCR with SARS-CoV-2 NP probes. All error bars indicate the standard errors of the mean. Mean values for the plaque size and half-life were compared using one-way ANOVA in GraphPad Prism 8.0 software. Statistical significance was set at p < 0.05.
Further studies are needed
Kwon and colleagues say it will be important to analyze these variants of concern in terms of both their clinical pathology and virology, as this will help to improve understanding of their increased virulence.
“For example, plaque size may be associated with contagiousness or viral transmission,” they write.
The researchers say that further studies are needed to identify additional determinants of plaque size, including functional mutations, interactions with host factors, and environmental composition.
“In addition, variants of interest and other variants should also be investigated to effectively control their spread,” concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Kwon YC, et al. Comparison of Plaque Size, Thermal Stability, and Replication Rate among SARS-CoV-2 Variants of Concern. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.09.30.462687, https://www.biorxiv.org/content/10.1101/2021.09.30.462687v1
- Peer reviewed and published scientific report.
Jeong, Gi Uk, Gun Young Yoon, Hyun Woo Moon, Wooseong Lee, Insu Hwang, Haesoo Kim, Kyun-Do Kim, et al. 2022. “Comparison of Plaque Size, Thermal Stability, and Replication Rate among SARS-CoV-2 Variants of Concern.” Viruses 14 (1): 55. https://doi.org/10.3390/v14010055. https://www.mdpi.com/1999-4915/14/1/55.