Coronavirus disease 19 (COVID-19), which is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is one of the five worldwide pandemics that has caused the highest fatality rates. COVID-19 is the third zoonotic coronavirus (CoV) pandemic that has occurred in the last 20 years; thus, this indicates that there are ample possibilities of future novel CoV outbreaks.
The high transmissibility of SARS-CoV-2, insufficient vaccinations, and the emergence of various SARS-CoV-2 variants suggest that this virus is highly likely to become an endemic and will therefore continue to pose a serious threat to humans. Effective antivirals that specifically target the most conserved viral enzymes would be an important mechanism for treatment against future novel CoVs.
At present, the most effective antivirals against COVID-19 are immune modulators that help to reduce the cytokine storm resulting from severe COVD-19. Presently remdesivir is the only antiviral that has been approved by the United States Food and Drug Administration (FDA) against SARS-CoV-2. Remdesivir is a repurposed ribonucleic acid (RNA)-dependent RNA polymerase (RdRp) inhibitor that is used for the treatment of COVID-19.
Two viral enzymes that can be targeted for in future effective antivirals are viral genome polymerases like RdRp, as well as virus-specific proteases that cleave the viral polyprotein into functional proteins. The efficacy of remdesivir has been found to be limited; however, remdesivir combined with a SARS-CoV-2 protease inhibitor could be highly effective against COVID-19.
SARS-CoV-2 encodes for two viral proteases, out of which CoV 3-chymotrypsin-like Main Protease (Mpro) is the more vital one. Mpro is involved in the generation of mature CoV non-structural proteins, including the RdRp subunits. Since Mpro is essential for the production of the viral genome replicase, it could be used for the development of a cell-based drug screening system.
A new study published in Antiviral Research aimed to report a cell-based assay that involved the isolation and quantification of SARS-CoV-2 Mpro in human cells in the absence of other proteins. The dose-response of drug activity against SARS-CoV-2 Mpro was evaluated, along with recovery of Mpro function after the drug was removed.
About the study
The current study involved HEK293 cells for determining the effect of drugs on the intracellular expression, as well as the function of Mpro. Specific deoxyribonucleic acid (DNA) plasmids were constructed for the expression of Mpro in these cells.
The total proteins were extracted from both transfected and untransfected cells that were either treated with drugs or mock-treated followed by immunoblotting. This was followed by fluorescence microscopy for quantification and MTT assay for determination of the cell viability. Furthermore, Calu-3 cells were used for SARS-CoV-2 live virus manipulations and RNA isolation.
Study findings
The results indicated that when the plasmid vector was expressed in HEK293 cells, it led to the production of a GST-Mpro fusion peptide. This fusion protein was found to self-cleave itself into GST and the fully functional Mpro protein.
The quantification of the protein was achieved through the use of a biosensor that involved the development of dimerization-dependent red fluorescent protein (RFP) with a caspase 3 cleavage site that was engineered between the two domains in a fusion protein construct. Cleavage of caspase 3 would result in loss of fluorescence.
The co-expression of Mpro with the biosensor resulted in cleavage of the biosensor and further led to the loss of RFP fluorescence. Furthermore, mutation of a glutamine residue in the Mpro cleavage site with alanine resulted in decreased cleavage of the biosensor and increased fluorescence as compared to wt Mro. Comparatively, mutation of the glutamine along with co-transfection with wt Mpro led to an even greater decrease in RFP cleavage and higher fluorescence.
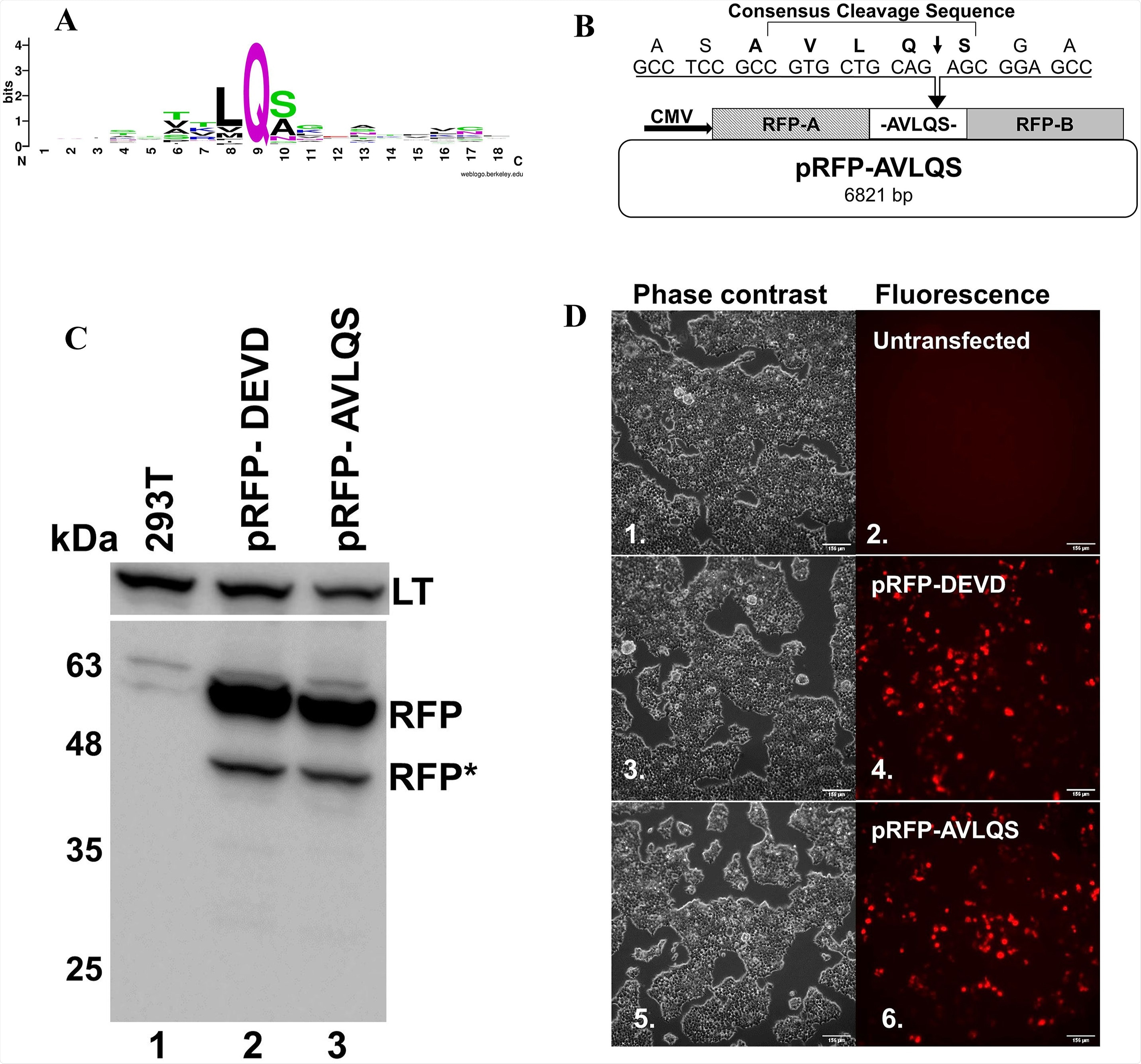 Creation and expression of an RFP biosensor to detect CoV2 Mprofunction: A) The consensus sequence for cleavage by Mpro, was computed using WebLogo version 3.0 using 77 cleavage sites, 11 each from SARS-CoV, MERS-CoV, SARS-CoV2, HCoV-229E, HCoV-NL63, HCoV-OC43 and HCoV-HKU1 (Supplemental Fig. 1). B) Graphic depiction of the pRFP-AVLQS vector, indicating the AVLQS cleavage site that was incorporated between the two RFP domains required to be tethered to allow for association and fluorescence. Expression of the RFP fusion protein is driven by the CMV promoter. C) Immunoblotting of HEK-293T cells untransfected (lane 1) or transfected with either the parental pRFPA1B1-DEVD vector (lane 2), or the pRFP-AVLQS vector (lane 3), for RFP (lower panel), or for SV40 Large T-antigen (LT, ∼94 kDa, upper panel) as a loading control. The heavy top band is the fluorescent RFP protein (RFP, ∼54 kDa) and the lower lighter band represents a breakdown product that is naturally produced upon expression in human cells (∼40 kDa RFP*) D) Phase contrast (odd) and fluorescence (even) images captured simultaneously using inverted microscopy demonstrate non-fluorescent untransfected cells (Cao et al., 2007), and similar levels of RFP fluorescence in cells transfected with either pRFPA1B1-DEVD or pRFP-AVLQS (compare 4 vs 6). Bar = 156 μm.
Creation and expression of an RFP biosensor to detect CoV2 Mprofunction: A) The consensus sequence for cleavage by Mpro, was computed using WebLogo version 3.0 using 77 cleavage sites, 11 each from SARS-CoV, MERS-CoV, SARS-CoV2, HCoV-229E, HCoV-NL63, HCoV-OC43 and HCoV-HKU1 (Supplemental Fig. 1). B) Graphic depiction of the pRFP-AVLQS vector, indicating the AVLQS cleavage site that was incorporated between the two RFP domains required to be tethered to allow for association and fluorescence. Expression of the RFP fusion protein is driven by the CMV promoter. C) Immunoblotting of HEK-293T cells untransfected (lane 1) or transfected with either the parental pRFPA1B1-DEVD vector (lane 2), or the pRFP-AVLQS vector (lane 3), for RFP (lower panel), or for SV40 Large T-antigen (LT, ∼94 kDa, upper panel) as a loading control. The heavy top band is the fluorescent RFP protein (RFP, ∼54 kDa) and the lower lighter band represents a breakdown product that is naturally produced upon expression in human cells (∼40 kDa RFP*) D) Phase contrast (odd) and fluorescence (even) images captured simultaneously using inverted microscopy demonstrate non-fluorescent untransfected cells (Cao et al., 2007), and similar levels of RFP fluorescence in cells transfected with either pRFPA1B1-DEVD or pRFP-AVLQS (compare 4 vs 6). Bar = 156 μm.
Evaluation of SARS-CoV-2 Mpro function took place with the help of a Mpro inhibitor known as GC376. Treatment of Mpro with ten-fold dilutions of 100 to 0.01 micromolar (µM) GC376 resulted in no cleavage of the biosensor, whereas treatment with 1 µM GC376 showed recovery of fluorescence levels. Thirdly, treatment with 10 and 100 µM GC376 showed high levels of RFP fluorescence recovery. No toxicity was observed following the treatment of cells with all concentrations of GC376.
The Mpro inhibitor activity of two FDA-approved drugs, Bepridil and Alverine, were compared to GC367. The results indicated that the extent of inhibition of intracellular Mpro function was higher with GC367 as compared to the two drugs. Furthermore, recovery of Mpro function was observed through the loss of fluorescence in cells expressing Mpro when the GC367 inhibitor was removed.
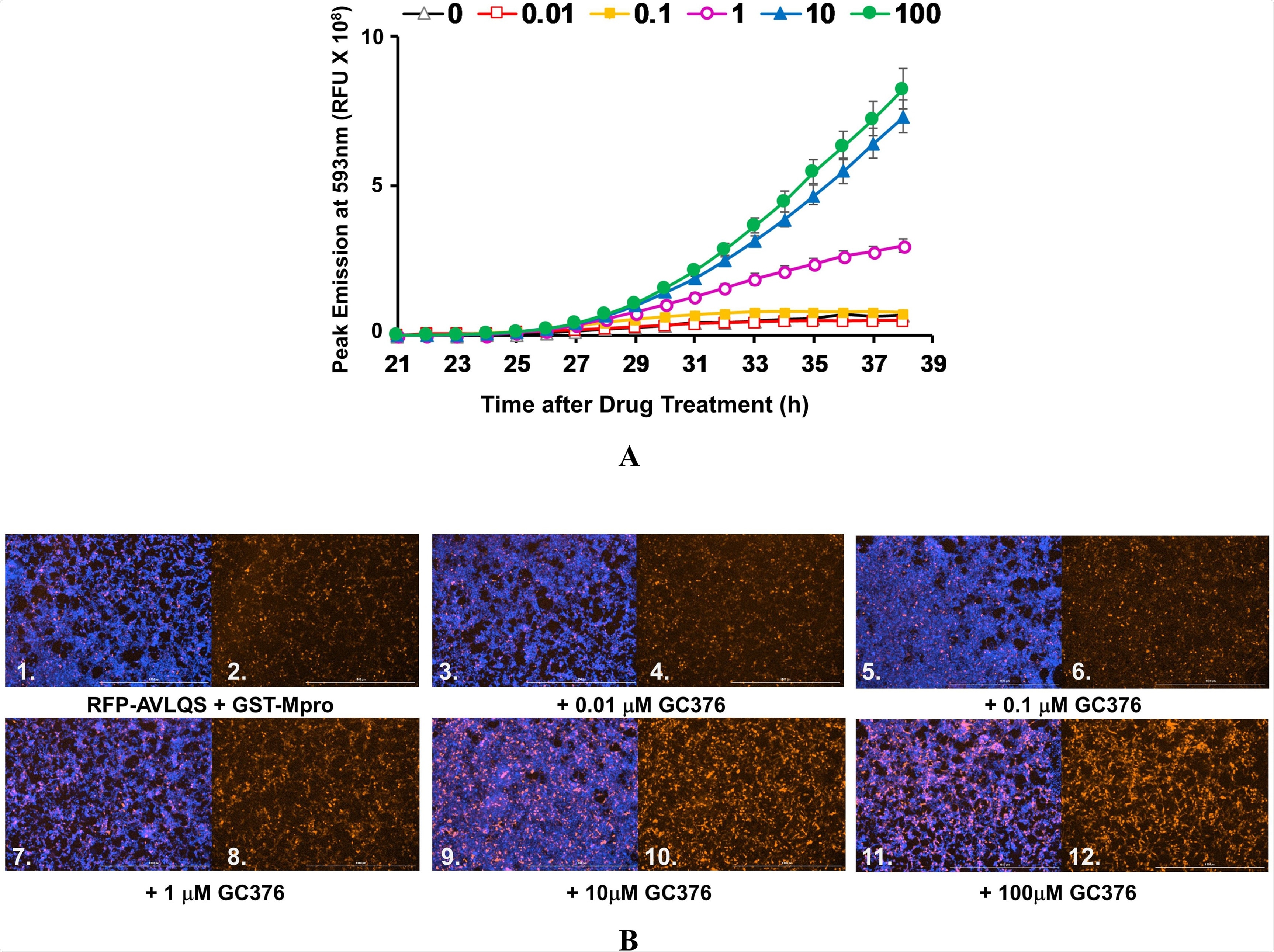 GC376 Inhibition of intra-cellular CoV2 Mpro: A) AD293 cells were co-transfected with pGST-Mpro and pRFP-AVLQS (ratio of 1:4) for 21 h. Cells stained with Hoechst were treated with varying levels of GC376: 0 μM (open triangles), 0.01 μM (open squares), 0.1 μM (filled squares), 1 μM (open circles), 10 μM (filled triangles), and 100 μM (filled circles). The fluorescence output of the live cells was monitored as in Fig. 3F for Hoechst-DNA and RFP every hour for 17 h following addition of GC376. Inhibition of Mpro by GC376 is indicated by increasing levels of fluorescence (note the greater sensitivity of the scale in Fig. 4A as compared to Fig. 3F). B) Images shown are from hour 36, or 15 h after addition of drug/OPTIMEM), visualizing for RFP (even panels) or RFP and DNA (odd panels), using the Cytation 1 imaging reader as described above. Bar = 1000 μm.
GC376 Inhibition of intra-cellular CoV2 Mpro: A) AD293 cells were co-transfected with pGST-Mpro and pRFP-AVLQS (ratio of 1:4) for 21 h. Cells stained with Hoechst were treated with varying levels of GC376: 0 μM (open triangles), 0.01 μM (open squares), 0.1 μM (filled squares), 1 μM (open circles), 10 μM (filled triangles), and 100 μM (filled circles). The fluorescence output of the live cells was monitored as in Fig. 3F for Hoechst-DNA and RFP every hour for 17 h following addition of GC376. Inhibition of Mpro by GC376 is indicated by increasing levels of fluorescence (note the greater sensitivity of the scale in Fig. 4A as compared to Fig. 3F). B) Images shown are from hour 36, or 15 h after addition of drug/OPTIMEM), visualizing for RFP (even panels) or RFP and DNA (odd panels), using the Cytation 1 imaging reader as described above. Bar = 1000 μm.
Conclusion
The current study was quite effective in determining the importance of Mpro for the development of antivirals. Although certain FDA-approved drugs having an affinity for intracellular Mpro and can therefore be repurposed for the treatment of COVID-19.
However, these drugs failed to show promising results. Therefore, the development of new antivirals with a higher affinity for Mpro is crucial, as these agents could inhibit their function and provide protection against SARS-CoV-2 and future zoonotic coronaviruses.