Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), responsible for coronavirus disease 2019 (COVID-19), continues to cause mayhem all over the world. Because of this, the scientific community has made rapid progress in understanding the molecular determinants of SARS-CoV-2 so that targeted therapies can be developed.
A critical stage in the SARS-CoV-2 life cycle is the replication-transcription machinery. These mechanisms are orchestrated by several nonstructural proteins like nsp12 with RNA-dependent RNA polymerase (RdRP), helicase (nsp13), an exonuclease (nsp14), an endonuclease (nsp15), and methyl-transferases (nsp14 and nsp16).
Amongst these, nsp13 plays an essential role in processive elongation by the RdRp on the highly structured RNA genome. However, this may also generate backtracked replication transcription machinery for proofreading and template-switching.
How these enzymes coordinate is yet to be explored; knowledge of these enzyme structures and dynamics can aid in gaining a comprehensive understanding of their roles.
Advanced techniques like cryo-electron microscopy have emerged as powerful tools in understanding large protein complexes and molecular machines.
The Study
With this background, the goal of a new study published on the bioRxiv* preprint server was to use cryo-electron microscopy to investigate the nsp13 helicase conformational states in order to gain insights into how nsp13 functions during viral transcription/replication.
The methodology of this study included the following steps:
.jpg)
Consensus cryo-EM structure of an nsp132-RTC. A. Overall architecture of the consensus nsp132-RTC. Shown is the transparent cryo-EM density (map3, local-resolution filtered) with the nsp132-RTC model superimposed. B. The consensus nsp132-RTC structure is shown; RNA is shown as atomic spheres, proteins are shown as transparent molecular surfaces. A low-pass filtered (6 Å) cryo-EM difference density reveals the path of the downstream t-RNA 5′-segment through the RNA binding groove of nsp13.1 (cyan surface).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The authors provided an in-depth structural analysis of a cryo-EM dataset and molecular dynamics (MD) simulation analysis of the nsp13-RTC (replication-transcription complex, or RTC), and the results can be segmented into seven sections.
In the first section, a series of cryo-EM maps were generated through focused refinement around sub-domains of the nsp131-RTC and nsp132-RTC (map3) maps. Then, these were combined to form a composite map with a nominal resolution of 2.8 Å.
A mask surrounding the nsp13.1 and nsp13.2 RecA1, RecA2, and 1B domains was then created. The masked classification with signal subtraction was used to identify four distinct conformational states with significant differences in the dispositions of the nsp13 subunits, particularly nsp13.1.
The authors reported that nsp13.1-engaged conformation grasps the downstream the RNA t-strand by interacting with the RNA phosphate backbone, which is mostly van der Waals interactions.
After that, a comparison was drawn between the nsp13.1-apo and nsp13.1-engaged structures. A striking change in the conformation of the RecA-like ATPase domains of nsp13.1 was reported. Furthermore, it was found that the conformation transition from the nsp13.1-engaged to the nsp13.1-apo structures corresponds to an ~21° rotation of the RecA2 domain concerning RecA1, opening the gap between the two domains.
The study revealed that in the nsp13.1-engaged state, the downstream single-stranded t-RNA is guided through a deep groove between the RecA1 and RecA2 domains that are entirely closed off whereas, in the 1B-open conformation, the nsp13.1 1B domain appears to be trapped open by the presence of nsp13.2. Thus, in this state, the single-stranded downstream t-RNA does not engage with the helicase and represents an inactive state of the helicase that would be unable to translocate on the RNA.
The authors combined the cryo-EM structural data with MD simulations. They found out that the conformation of the nsp13.1 1B domain in the 1B-open structure is not stable on its own but is sterically trapped by the presence of nsp13.2, blocking the conformational change required for 1B domain closure.
The MD simulation studies also helped authors understand the backtracking on and off (proofreading) mechanism of nsp132-RTC that turns on, using an allosteric mechanism to switch between RNA synthesis or backtracking in response to stimuli at the RdRp active site.
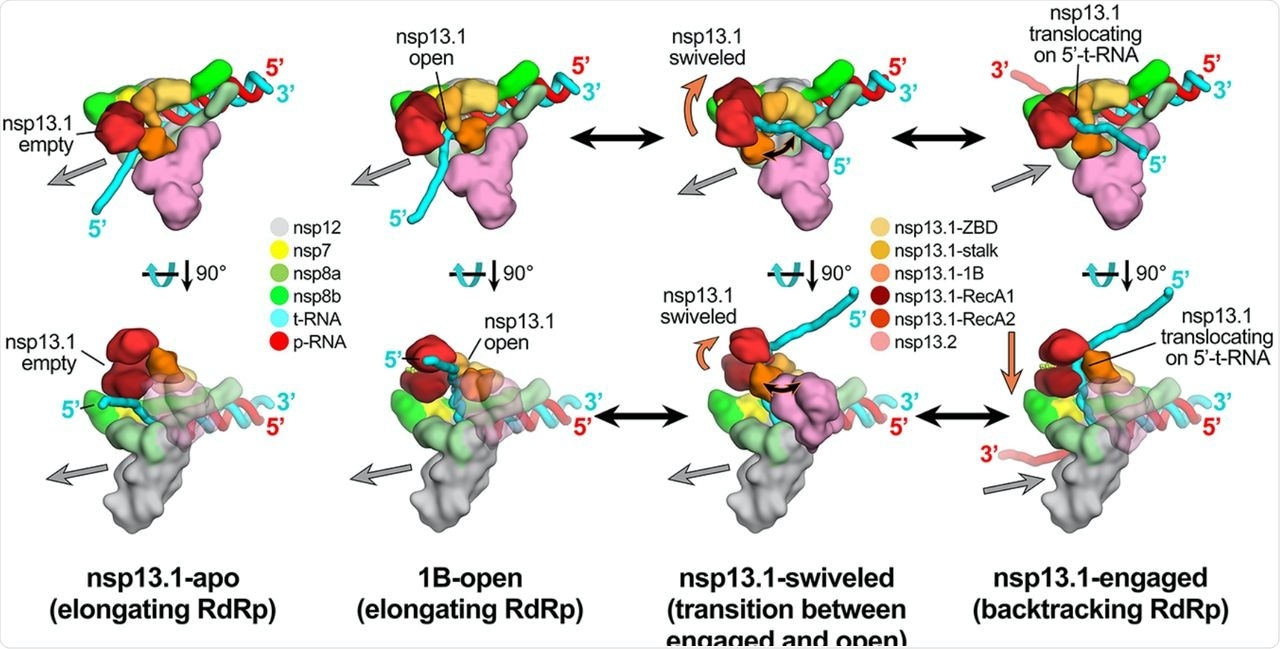
Schematic model for RTC elongation (1B-open) vs. backtracking (nsp13.1-engaged) states. Top views (top row) and side views (bottom row) of each structural class. Nsp13.1-apo (17%): The nsp13.1 RecA domains are open, consistent with the absence of nucleotide. The nsp13.1 is therefore not engaged with the downstream 5′-t-RNA and the RdRp can freely translocate on the t-RNA with concurrent elongation of the p-RNA (gray arrow pointing downstream). 1B-open (33%): The nsp13.1 1B domain is rotated open and sterically trapped by the presence of nsp13.2. The nsp13.1 is therefore unable to engage with the downstream 5′-t-RNA and is inactive. The RdRp is able to elongate freely in the downstream direction. Nsp13.1-swiveled (17%): The rotation of the nsp13.1 protomer away from nsp13.2 provides space for the nsp13.1 1B domain to open and/or close. We therefore propose that nsp13.1-swiveled represents a transition state between the 1B-open (elongating) and nsp13.1-engaged (backtracking) states. Nsp13.1-engaged (33%): The nsp13.1 1B and RecA domains are clamped onto the downstream 5′-t-RNA. In this state, nsp13.1 can translocate on the t-RNA in the 5′-3′ direction (shown by the orange arrow). This counteracts RdRp elongation and causes backtracking (backward motion of the RdRp on the RNA, shown by the gray arrow pointing upstream).
Conclusion
These findings revealed that distinct conformational states of the nsp13 protomers within the SARS-CoV-2 nsp132-RTC, shedding light on the function of nsp13 and its complex with the RTC.
Furthermore, it was suggested that nsp13 backtracking would allow for nsp10/nsp14 3'-exonuclease proofreading activity to access and degrade the mismatched p-RNA 3'-nucleotide. Thus, the virus's nsp14-mediated proofreading activity is critical for avoiding a mutation catastrophe during replication and is also a key determinant of SARS-CoV-2 susceptibility to many anti-viral nucleotide analogs.
An identical analysis of the backtracked nsp132-BTC revealed a remarkably different distribution of particles in which the nsp13.1-engaged state was heavily favored. From this, it was speculated that the conformational switch that turns to backtrack on and off is allosterically controlled.
It must be noted that no statistical methods were used to determine sample size, the experiments were not randomized, and the investigators were not blinded during the experiments and outcome assessment.
The strength of this study lies in its extensive and robust structural analysis of a cryo-EM dataset of the nsp13-RTC, combined with MD simulation analysis of the resulting structures.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Chen, J., Wang, Q., Malone, B., et al. (2021), “Ensemble cryo-electron microscopy reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex”, bioRxiv, doi: 10.1101/2021.11.10.468168, https://www.biorxiv.org/content/10.1101/2021.11.10.468168v1
- Peer reviewed and published scientific report.
Chen, James, Qi Wang, Brandon Malone, Eliza Llewellyn, Yakov Pechersky, Kashyap Maruthi, Ed T. Eng, et al. 2022. “Ensemble Cryo-EM Reveals Conformational States of the Nsp13 Helicase in the SARS-CoV-2 Helicase Replication–Transcription Complex.” Nature Structural & Molecular Biology 29 (3): 250–60. https://doi.org/10.1038/s41594-022-00734-6. https://www.nature.com/articles/s41594-022-00734-6.