Natural immunity plays a critical role in protecting us from the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the pathogen responsible for coronavirus disease 2019 (COVID-19).
Interferons (IFNs) constitute a significant part of the anti-viral innate defense system. To establish an infection, SARS-CoV-2 needs to cross this defense mechanism of interferons.
The molecular patterns of viral pathogens are recognized by pattern recognition receptors which leads to the activation of signal cascades. This causes the induction and secretion of IFNs as well as other pro-inflammatory cytokines.
Further, these secreted IFNs bind to their respective receptors, which leads to the induction and expression of several interferons stimulated genes (ISGs).
Interferons are categorized into three types: Human type I IFNs (includes IFNα and IFNβ), Type II IFNs (includes IFNγ), and Type III IFN (includes IFNλ1-4).
Due to the continuous adaptation of SARS-CoV-2 to the human population, multiple variants of concerns (VOCs) have emerged with increased infectiousness and decreased sensitivity to preventive or therapeutic measures.
The World Health Organization (WHO) has classified four SARS-CoV-2 variants as VOCs: B.1.1.7, B.1.351, P.1, and B.1.617.2. These are also known as alpha, beta, gamma, and delta variants, respectively.
Comparing the replication efficiency of SARS-CoV-2 variants of concerns with the early isolate of SARS-CoV-2
In a study recently published on the bioRxiv* preprint server, researchers investigated the difference in replication efficiency or IFN sensitivity of the four VOCs.
 Study: The Delta variant of SARS-CoV-2 maintains high sensitivity to interferons in human lung cells. Image Credit: NIAID
Study: The Delta variant of SARS-CoV-2 maintains high sensitivity to interferons in human lung cells. Image Credit: NIAID

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Since the first available sequence of the Wuhan-Hu-1 isolate, mutations were observed in the S glycoprotein, proteins involved in replication, and innate immune escape of SARS-CoV-2 isolate from February 2020 (NL-02- 2020) and four VOC isolates.
In this study, the researchers utilized the human epithelial lung cancer cell line Calu-3 and found that as compared to the NL-02-2020 isolate, the Delta variant replicated with greater efficiency.
Also, the SARS-CoV-2 alpha and gamma variants depicted intermediate phenotypes, whereas the SARS-CoV-2 beta variant replicated with moderately reduced efficiency compared to NL-02-2020.
The iPSC-derived alveolar epithelial type II (iAT2) cells comprise a major part of pulmonary alveolar epithelial cells and are considered the main targets of SARS-CoV-2 in the distal lung.
The researchers observed that compared to other SARS-CoV-2 isolates, the SARS-CoV-2 beta variant replicated with moderately reduced efficiency in the iAT2 cells.
With the increase in virus-induced loss of AT2 cells, the severity of COVID-19 also increases. The results showed that the sensitivity of NL-02-2020 isolate was more towards IFNβ, IFNγ, and IFNλ1 than toward IFNα2 in Calu-3 cells.
Also, it was demonstrated that in the presence of different concentrations of IFNs, the SARS-CoV-2 alpha variant showed the highest resistance against IFN treatment.
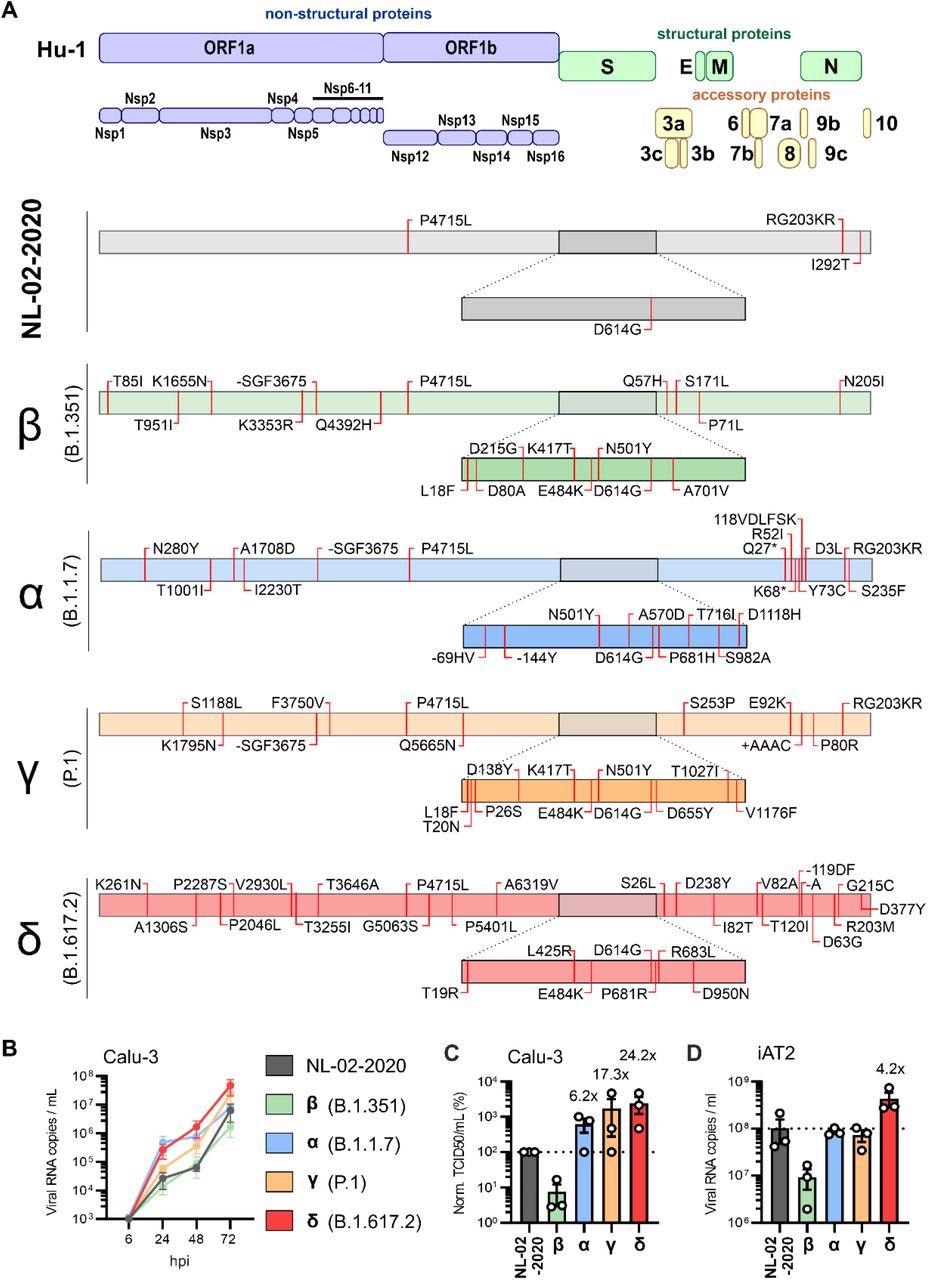 Amino acid differences and replication kinetics of early SARS-CoV-2 and VOCs. A, Schematic depiction of the SARS-CoV-2 genomic arrangement and proteins (top). Outline of the specific amino acid exchanges compared to the reference Hu-1 sequence in an early European Feb 2020 SARS-CoV-2 isolate (NL-02-2020), and four variants of concern in the order of appearance: Beta (B.1.351), Alpha (B.1.1.7), Gamma (P.1) and Delta (B.1.617.2) as assessed by next-generation sequencing assembly of the full genome. B, Viral RNA in the supernatant of Calu-3 cells infected with indicated SARS-CoV-2 variants was quantified by qRT-PCR at indicated timepoints post infection (MOI 0.05). Day 0 wash CTRL values were subtracted from data shown in the panel. n=3±SEM. C, Infectious SARS-CoV-2 particles in the supernatant, corresponding to the 72h post-infection time point shown in (B). n=3±SEM. D, Viral RNA in the supernatant of iAT2 cells infected with indicated SARS-CoV-2 variants was quantified by qRT-PCR at 2 days post infection (MOI 0.5) day 0 wash control values were subtracted from data shown in the panel. n=3±SEM.
Amino acid differences and replication kinetics of early SARS-CoV-2 and VOCs. A, Schematic depiction of the SARS-CoV-2 genomic arrangement and proteins (top). Outline of the specific amino acid exchanges compared to the reference Hu-1 sequence in an early European Feb 2020 SARS-CoV-2 isolate (NL-02-2020), and four variants of concern in the order of appearance: Beta (B.1.351), Alpha (B.1.1.7), Gamma (P.1) and Delta (B.1.617.2) as assessed by next-generation sequencing assembly of the full genome. B, Viral RNA in the supernatant of Calu-3 cells infected with indicated SARS-CoV-2 variants was quantified by qRT-PCR at indicated timepoints post infection (MOI 0.05). Day 0 wash CTRL values were subtracted from data shown in the panel. n=3±SEM. C, Infectious SARS-CoV-2 particles in the supernatant, corresponding to the 72h post-infection time point shown in (B). n=3±SEM. D, Viral RNA in the supernatant of iAT2 cells infected with indicated SARS-CoV-2 variants was quantified by qRT-PCR at 2 days post infection (MOI 0.5) day 0 wash control values were subtracted from data shown in the panel. n=3±SEM.
The SARS-CoV-2 delta variant was at least as sensitive towards IFN treatment as the NL-02-2020 isolate.
Furthermore, as compared in Calu-3 cells, type II IFN was less effective against SARS-CoV-2 in iAT2 cells. The SARS-CoV-2 alpha variant was the least vulnerable to IFNs, and the SARS-CoV-2 alpha, beta, and gamma variants show higher resistance against type III IFN. The SARS-CoV-2 delta variant remains as sensitive to IFNs as early 2020 SARS-CoV-2 isolates.
“The Alpha variant shows reduced susceptibility to IFNs.”
Findings could help improve IFN therapies against SARS-CoV-2 delta VOC
The emergence of SARS-CoV-2 VOCs could be attributed to various features like replication efficiency, the efficiency of immune evasion, and virion infectivity.
The combination of the Alpha variant’s IFN resistance and the increased replication fitness of the Delta variant poses an increased risk for the emergence of a new VOC which can be more harmful than existing VOCs.
The deletion in Nsp6 has been found to be a shared trait among SARS-CoV-2 alpha and gamma VOCs with reduced sensitivity to type III IFN.
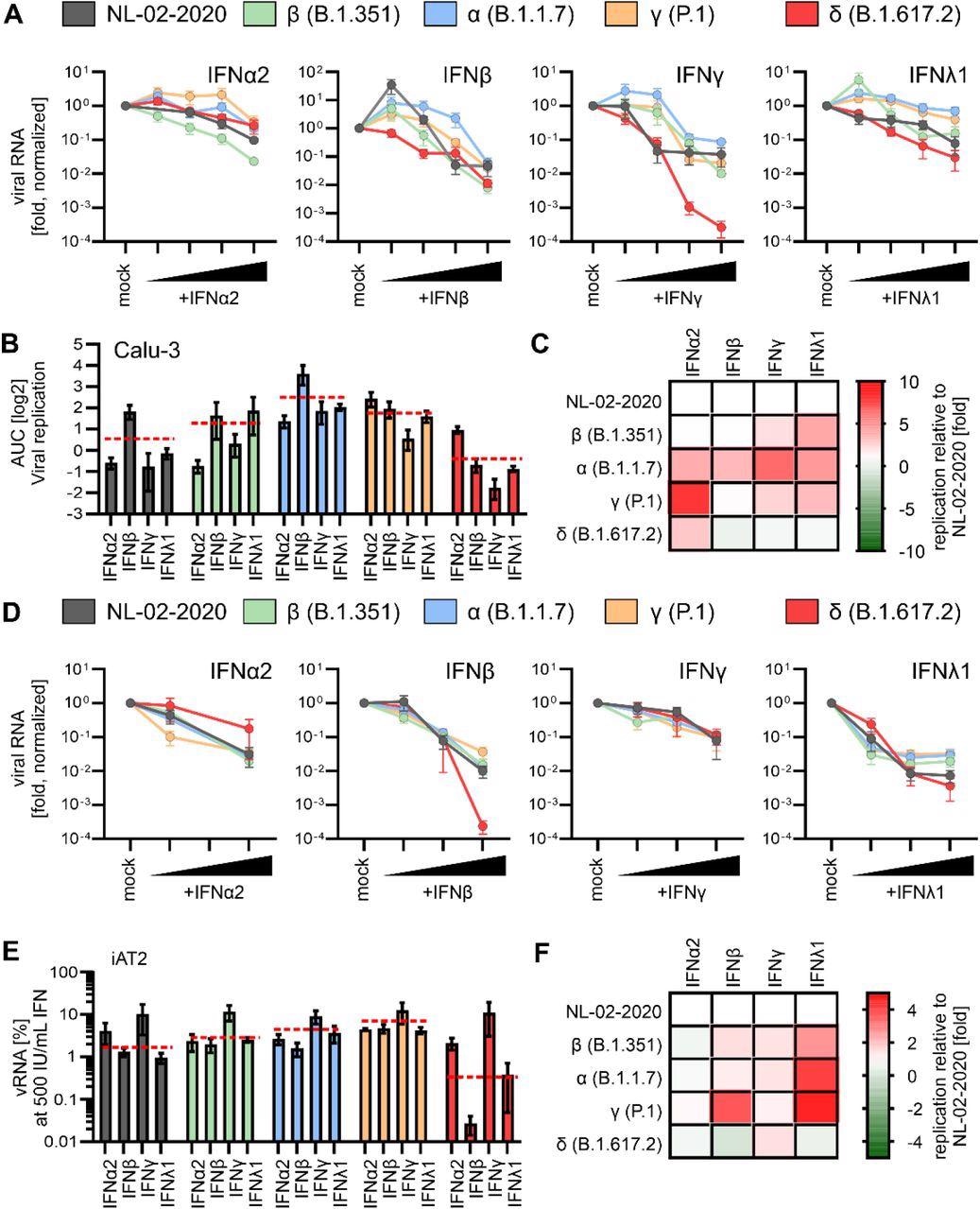 Interferon sensitivity of NL-02-2020 and VOCs. A, Normalized amount of viral RNA in the supernatant of Calu-3 cells infected with indicated SARS-CoV-2 variants was quantified by qRT-PCR at 72h post-infection (MOI 0.05, no IFN set to 100%). Cells were infected 3 days post treatment with indicated IFNs (α2, β and γ 0.5, 5, 50 and 500 U/ml) or IFNλ1 (0.1, 1, 10 and 100 ng/ml). n=2±SEM. B, Area under the curve analysis of the data in (A) representing the replication of the variants in the presence of IFNs. Red lines indicate the average over all IFNs for one variant. C, Heatmap displaying differences in viral replication (Area under the curve analysis) of the VOCs compared to the NL-02-2020 variant upon IFN treatment of Calu-3 cells. Red, increased replication, green, decreased replication relative to replication of NL-02-2020. Data from (A). D, Normalized amount of viral RNA in the supernatant of iAT2 cells infected with indicated SARS-CoV-2 variants as quantified by qRT-PCR at 48h post-infection (MOI 0.5, no IFN set to 100%). Cells were infected for 2 days post treatment with indicated IFNs (α2, β and γ: 5, 50 and 500 U/ml) or IFNλ1 (1, 10 and 100 ng/ml). n=4±SEM. E, Percentage of viral RNA in the supernatant of iAT2 cells as a fraction between non-treated and IFN treated (500 IU/mL or 100 ng/mL). data from (D). Red lines indicate the average over all IFNs for one variant. F, Heatmap displaying fold differences in viral replication of the VOCs in iAT2 cells compared to the NL-02-2020 variant (set to 1) upon treatment with IFN (500 IU/mL or 100 ng/mL). Red, increased replication, green, decreased replication relative to NL-02-2020. Data from (D).
Interferon sensitivity of NL-02-2020 and VOCs. A, Normalized amount of viral RNA in the supernatant of Calu-3 cells infected with indicated SARS-CoV-2 variants was quantified by qRT-PCR at 72h post-infection (MOI 0.05, no IFN set to 100%). Cells were infected 3 days post treatment with indicated IFNs (α2, β and γ 0.5, 5, 50 and 500 U/ml) or IFNλ1 (0.1, 1, 10 and 100 ng/ml). n=2±SEM. B, Area under the curve analysis of the data in (A) representing the replication of the variants in the presence of IFNs. Red lines indicate the average over all IFNs for one variant. C, Heatmap displaying differences in viral replication (Area under the curve analysis) of the VOCs compared to the NL-02-2020 variant upon IFN treatment of Calu-3 cells. Red, increased replication, green, decreased replication relative to replication of NL-02-2020. Data from (A). D, Normalized amount of viral RNA in the supernatant of iAT2 cells infected with indicated SARS-CoV-2 variants as quantified by qRT-PCR at 48h post-infection (MOI 0.5, no IFN set to 100%). Cells were infected for 2 days post treatment with indicated IFNs (α2, β and γ: 5, 50 and 500 U/ml) or IFNλ1 (1, 10 and 100 ng/ml). n=4±SEM. E, Percentage of viral RNA in the supernatant of iAT2 cells as a fraction between non-treated and IFN treated (500 IU/mL or 100 ng/mL). data from (D). Red lines indicate the average over all IFNs for one variant. F, Heatmap displaying fold differences in viral replication of the VOCs in iAT2 cells compared to the NL-02-2020 variant (set to 1) upon treatment with IFN (500 IU/mL or 100 ng/mL). Red, increased replication, green, decreased replication relative to NL-02-2020. Data from (D).
The study results show that IFNβ and IFNλ1 effectively inhibit the SARS-CoV-2 delta variant in human alveolar epithelial type II cells. This can help improve IFN-based therapies against the currently dominant SARS-CoV-2 delta VOC.
“Further studies on the molecular determinants of reduced IFN sensitivity and improved innate immune evasion of emerging SARS-CoV-2 variants are highly warranted.”

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference:
- Preliminary scientific report.
The Delta variant of SARS-CoV-2 maintains high sensitivity to interferons in human lung cells. Rayhane Nchioua, Annika Schundner, Dorota Kmiec, Caterina Prelli Bozzo, Fabian Zech, Lennart Koepke, Manfred Frick, Konstantin M. J. Sparrer, Frank Kirchhoff, bioRxiv, 2021. DOI: https://doi.org/10.1101/2021.11.17.468942, https://www.biorxiv.org/content/10.1101/2021.11.16.468777v1