T cell receptor (TCR) repertoires are surface proteins uniquely expressed by T cells. An effective T-cell response can help clear respiratory viral infections, and having T-cells with diverse TCR repertoires can help identify a wide variety of pathogens.
Understanding the changes in TCR repertoires during viral infection may help uncover more information on T cell adaptive immune response against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
In addition, unlike neutralizing antibodies that target the viral surface, T cells cover a wider distribution of targets, making it harder for SARS-CoV-2 variants to evade. Therefore, vaccination strategies that target the spike protein and stimulate T-cell response may provide broader and more robust protection.
 Study: T cell receptor repertoire signatures associated with COVID-19 severity. Image Credit: fusebulb / Shutterstock
Study: T cell receptor repertoire signatures associated with COVID-19 severity. Image Credit: fusebulb / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The study
The researchers created an integrative, systems immunology approach to study TCR repertoires and, thereby, the adaptive immune response of patients with different severity levels in COVID-19 infection and patients without COVID-19.
The investigation into TCR specificity groups for COVID-19 included over 4 billion clones across 2,130 repertoires.
The researchers sequencing TCR data found that COVID-19 infection shifted the immune repertoire. Patients with COVID-19 had reduced diversity of repertoires and reshaped clonal representation. Additionally, only a small expansion of functional clones was available during viral infection.
Further analysis found differences in CDR3 sequences from patient data collected across multiple cohorts. CDR3 sequences are a region on the immunoglobulin heavy chain that can influence how antibodies identify a target and bind.
CDR3 sequences found in mild, moderate, and severe disease had considerable overlap, while there was little similarity in CDR3 sequences of healthy patients. Across all samples of patients infected with COVID-19, the researchers found 42 conserved CDR3 sequences.
The researchers noted that sequence convergence in immune repertoires might also occur on a lower level, such as sequence substrings called motifs. They decomposed CDR3 sequences into motifs by overlapping amino acid sequences of length k (k-mers).
After creating 3 to 6 k-mers, they grouped sequences by samples with varying disease severity. Results showed that samples of patients with severe disease clustered together but were separate from samples with mild to moderate infection.
The results confirm clinical findings that patients with severe COVID-19 infection tend to have significant immune dysregulation than those with minor to moderate infection. Additionally, CDR3 sequencing may help with distinguishing patients with severe COVID-19 illness.
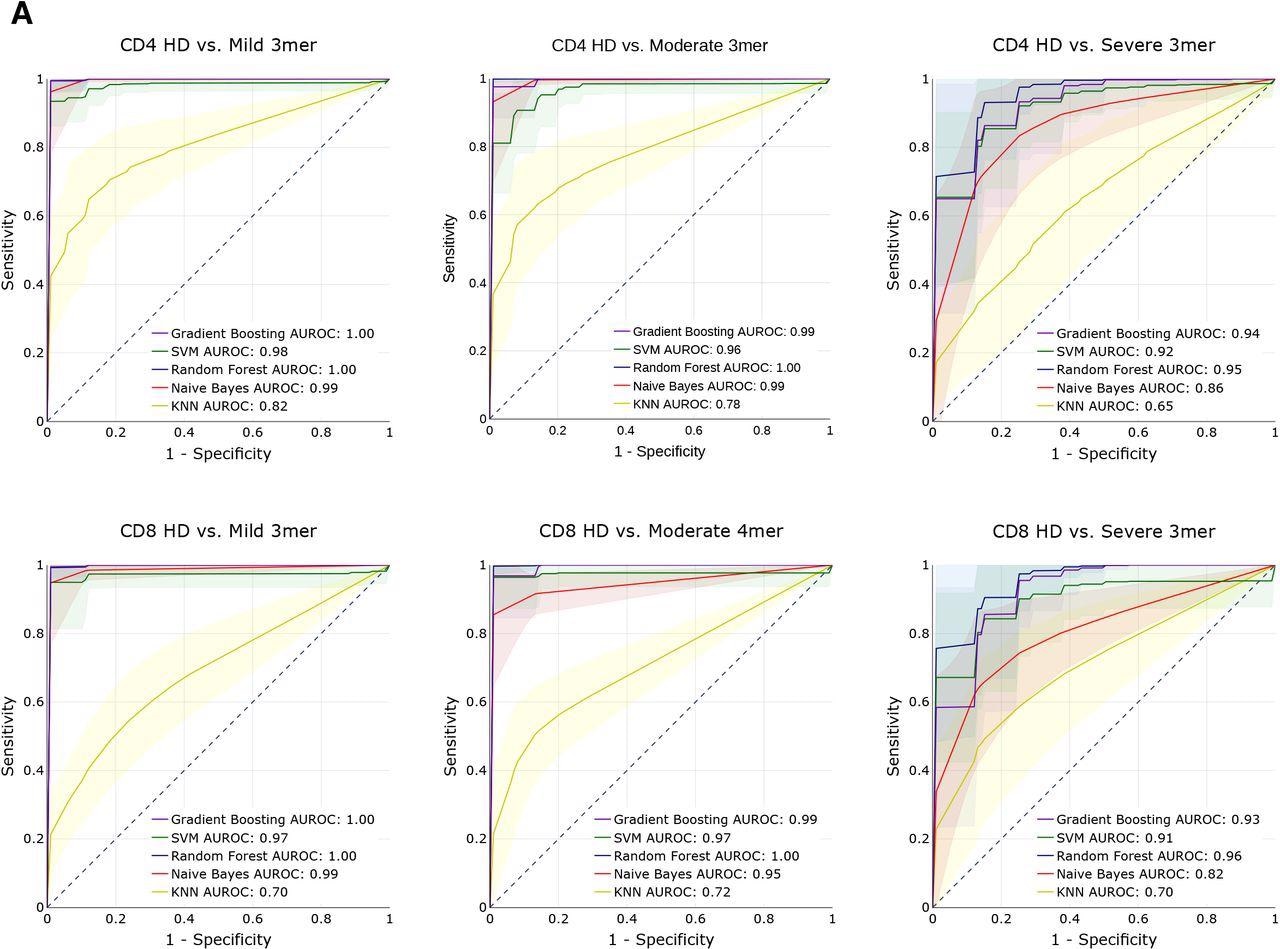 Predictive performance of machine learning models for disease severity. (A) AUROC curves for five machine learning models (gradient boosting trees, support vector machines, random forests, Naïve Bayes, and k-nearest neighbors) using 3-mer representations of TCR repertoire data. Models were trained to predict disease severity (mild, moderate, severe) vs healthy donors for CD4 (top row) and CD8 (bottom row) samples. Training and evaluation was performed using 100 repetitions of 5-fold cross-validations per model, average performance +/− 1 standard deviation shown on individual plots.
Predictive performance of machine learning models for disease severity. (A) AUROC curves for five machine learning models (gradient boosting trees, support vector machines, random forests, Naïve Bayes, and k-nearest neighbors) using 3-mer representations of TCR repertoire data. Models were trained to predict disease severity (mild, moderate, severe) vs healthy donors for CD4 (top row) and CD8 (bottom row) samples. Training and evaluation was performed using 100 repetitions of 5-fold cross-validations per model, average performance +/− 1 standard deviation shown on individual plots.
Heatmaps also showed shared motifs, including YNE, NEQ, EQF, and QFF, for repertoires of mild and moderate samples.
Using two other sequence similarity approaches, the researchers also looked at TCR-repertoire specificity clusters as they could help predict antigen specificity.
About 677 clusters were associated with disease, and 474 were exclusive to COVID-19 infection. There was no disease-associated cluster that overlapped with clusters from healthy patients.
Next, the researchers looked at gene rearrangements in TCR genes and disease-associated features such as clonal expansion. They used T cell single-cell RNA sequencing to study samples with CDR3 sequences associated with GLIPH2 clusters.
There were 12 clusters with different gene signatures. In cluster 6, cells had the greatest degree of clonality. A correlation was also found between clonality and disease severity.
Samples from patients with COVID-19 infection showed the greatest density in effector phenotype associated cluster 6. In contrast, cells from healthy patient samples had density in the naive phenotype-associated clusters.
A high density of cells from samples with COVID-19 was observed in clusters with high degrees of clonality. Meanwhile, there was a low density of disease-associated cells in clusters with low degrees of clonality.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources