In a recent study conducted at Yale University, USA, scientists have investigated the immunogenicity of mRNA-based coronavirus disease 2019 (COVID-19) vaccine candidates encoding the spike proteins of different severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants. The findings reveal that the vaccines induce robust immune responses against SARS-CoV-2 variants. The study is currently available on the bioRxiv* preprint server.
 Study: Systems immune profiling of variant-specific vaccination against SARS-CoV-2. Image Credit: NIAID
Study: Systems immune profiling of variant-specific vaccination against SARS-CoV-2. Image Credit: NIAID

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
The continuous emergence of novel SARS-CoV-2 variants with multiple spike mutations has caused a significant reduction in vaccine efficacy worldwide. For instance, the beta variant of SARS-CoV-2, which was first detected in South Africa, has caused a decrease in the effectiveness of the mRNA-based Pfizer/BioNTech COVID-19 vaccine from 90% to 70% by acquiring escape mutations. A similar effect has been observed for the delta variant, which was detected first in India. In addition, there is also grave concern about the recent arrival of the new Omicron variant.
Given the significant negative impacts of viral variants on vaccine efficacy, it is essential to develop next-generation vaccines that can directly target and neutralize more deadly variants of SARS-CoV-2.
In the current study, the scientists have developed three lipid nanoparticle-formulated mRNA-based vaccine candidates that encode the spike proteins of wild-type SARS-CoV-2 and its variants B.1.351 (Beta) and B.1.617 (14 lineages, including B.1.617.1 “Kappa variant,” B.1.617.2 “Delta variant). In addition, they have specifically investigated the immunogenicity of these vaccine candidates in mice.
They immunized the mice with two doses (prime-boost) of each vaccine intramuscularly at an interval of three weeks. They collected serum samples from the mice two weeks after vaccination to assess antibody response. The mice were sacrificed after 40 days of vaccination and their spleens, lymph nodes, and blood cells were collected for flow cytometry, single-cell transcriptomics, B cell receptor sequencing, and T cell receptor sequencing.
Immune responses to wild-type vaccine
The immunization of mice with wild-type vaccine resulted in strong binding and neutralizing antibody responses against the wild-type virus and all tested variants (B.1.351 and B.1.617). However, the neutralizing potency of vaccinated sera was many folds lower against the variants compared to that against the wild-type virus.
Regarding cellular immunity, the wild-type vaccine showed high efficacy in inducing spike-specific CD8+ T cells secreting interferon-gamma, tumor necrosis factor-alpha, and interleukin-2. Similarly, the vaccine-induced robust spike-specific CD4+ T Cells secreting interferon-gamma.
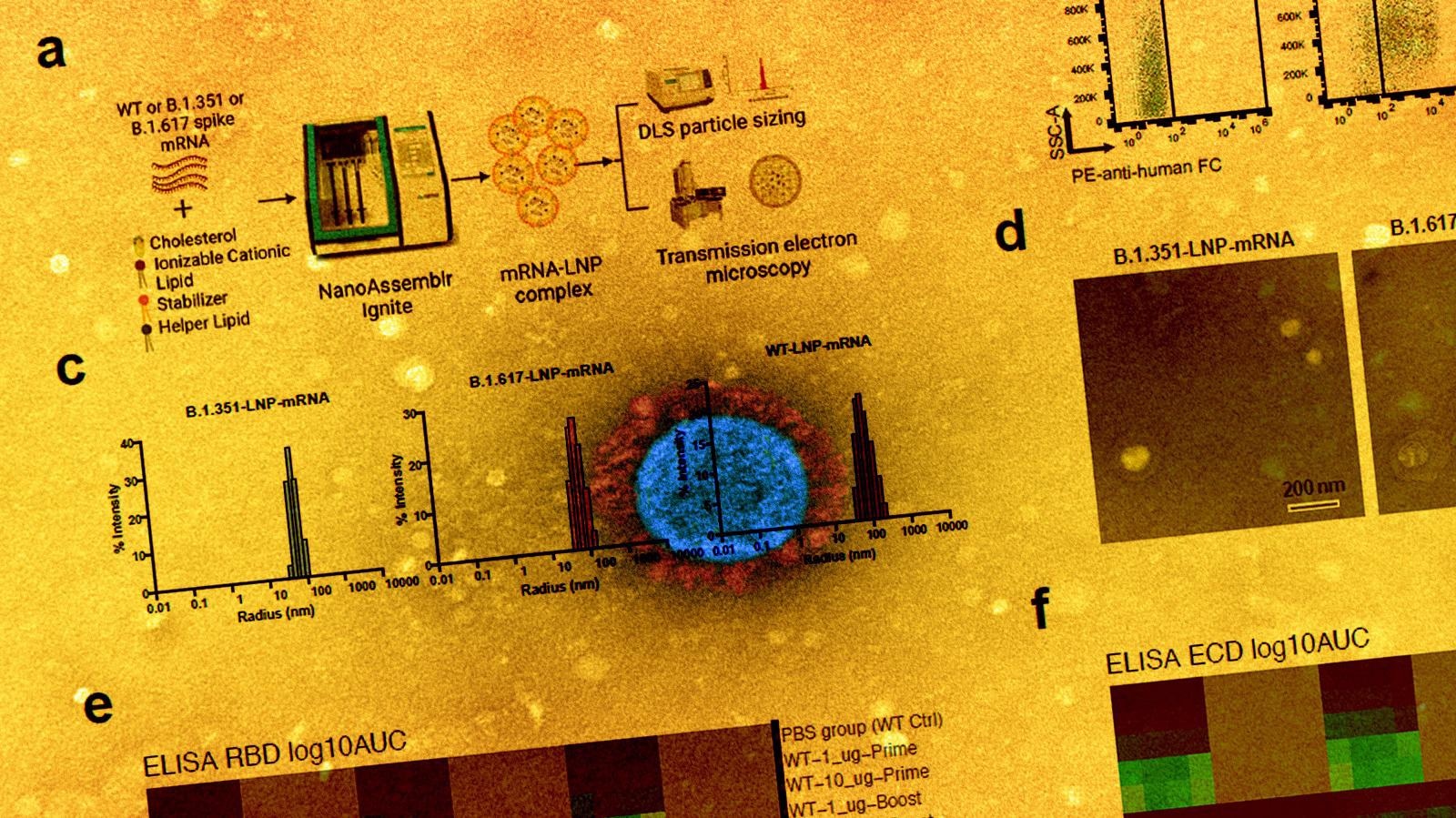
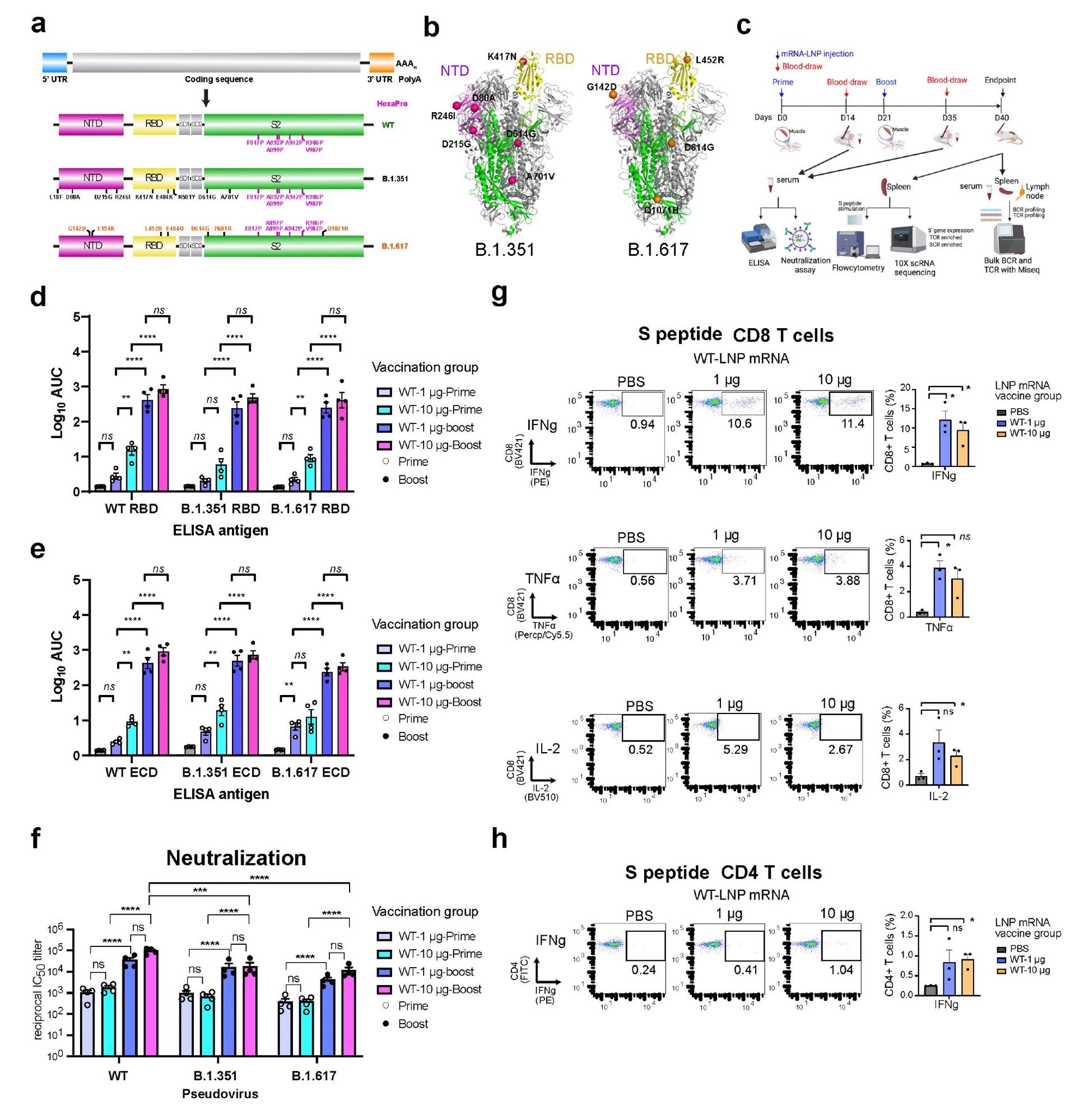 Overview of the primary experimental design, and the B and T cell responses induced by WT3 LNP-mRNA vaccination against SARS-CoV-2 WT, B.1.351 and B.1.617 spikes in mice. a, Schematic of the designs of three variant-specific LNP-mRNA vaccine candidates. Functional elements were shown in the spike mRNA and translated protein of SARS-CoV-2 WT, B.1.351 and B.1.617 spikes, including protein domains, HexaPro and variant-specific mutations. b, 3D structure highlighting certain variant-specific mutations in B.1.351 and B.1.617 spikes. The distribution of mutations of B.1.351 and B.1.617 was shown in the structure of SARS-CoV-2 (PDB: 6VSB). Mutations of B.1.351 and B.1.617 were shown as spheres, except for those in the unstructured loop regions. Certain mutations were not visible in the structure as they fall into floppy regions of the spike. c, Schematic of the overall design of primary experiments. Six- to 8-week-old C57BL/6Ncr mice (B.1.351-LNP12 mRNA (top) and B.1.617-LNP-mRNA, n = 6 mice per group; WT-LNP-mRNA, n = 4 mice; PBS, n = 9) received 1 or 10 µg of WT-LNP mRNA, B.1.351-LNP-mRNA or B.1.617-LNP-mRNA via the intramuscular route on day 0 (Prime) and day 21 (Boost). Blood was collected twice, two weeks post-prime and boost. The binding and pseudovirus-neutralizing antibody responses induced by LNP-mRNA were evaluated by ELISA and neutralization assay. Mice were euthanized at day 40. The spleen, lymph node, and blood samples were collected to analyze immune responses in by flow cytometry, bulk BCR and TCR profiling and single-cell profiling. d-e, Serum ELISA titers of WT-LNP mRNA vaccinated animals (n = 4). Serum antibody titer as the area under the curve (AUC) of the log10-transformed curve (1og10 AUC) to spike RBDs (d) and ECDs (e) of SARS-CoV-2 WT, B.1.351 and B.1.617. Two-way ANOVA with Tukey's multiple comparisons test was used to assess statistical significance. f, Serum neutralization titers of WT-LNP mRNA vaccinated animals (n = 4). Cross neutralization of SARS-CoV-2 WT, B.1.351 or B.1.617 pseudovirus infection of ACE2-overexpressed 293T cells. Two-way ANOVA with Tukey's multiple comparisons test was used to assess statistical significance. g-h, T cell response of WT-LNP mRNA vaccinated animals (n = 4). CD8+ (g) and CD4+ (h) T cell responses were measured by intracellular cytokine staining 6 hours after the addition of BFA. The unpaired parametric t-test was used to evaluate the statistical significance.
Overview of the primary experimental design, and the B and T cell responses induced by WT3 LNP-mRNA vaccination against SARS-CoV-2 WT, B.1.351 and B.1.617 spikes in mice. a, Schematic of the designs of three variant-specific LNP-mRNA vaccine candidates. Functional elements were shown in the spike mRNA and translated protein of SARS-CoV-2 WT, B.1.351 and B.1.617 spikes, including protein domains, HexaPro and variant-specific mutations. b, 3D structure highlighting certain variant-specific mutations in B.1.351 and B.1.617 spikes. The distribution of mutations of B.1.351 and B.1.617 was shown in the structure of SARS-CoV-2 (PDB: 6VSB). Mutations of B.1.351 and B.1.617 were shown as spheres, except for those in the unstructured loop regions. Certain mutations were not visible in the structure as they fall into floppy regions of the spike. c, Schematic of the overall design of primary experiments. Six- to 8-week-old C57BL/6Ncr mice (B.1.351-LNP12 mRNA (top) and B.1.617-LNP-mRNA, n = 6 mice per group; WT-LNP-mRNA, n = 4 mice; PBS, n = 9) received 1 or 10 µg of WT-LNP mRNA, B.1.351-LNP-mRNA or B.1.617-LNP-mRNA via the intramuscular route on day 0 (Prime) and day 21 (Boost). Blood was collected twice, two weeks post-prime and boost. The binding and pseudovirus-neutralizing antibody responses induced by LNP-mRNA were evaluated by ELISA and neutralization assay. Mice were euthanized at day 40. The spleen, lymph node, and blood samples were collected to analyze immune responses in by flow cytometry, bulk BCR and TCR profiling and single-cell profiling. d-e, Serum ELISA titers of WT-LNP mRNA vaccinated animals (n = 4). Serum antibody titer as the area under the curve (AUC) of the log10-transformed curve (1og10 AUC) to spike RBDs (d) and ECDs (e) of SARS-CoV-2 WT, B.1.351 and B.1.617. Two-way ANOVA with Tukey's multiple comparisons test was used to assess statistical significance. f, Serum neutralization titers of WT-LNP mRNA vaccinated animals (n = 4). Cross neutralization of SARS-CoV-2 WT, B.1.351 or B.1.617 pseudovirus infection of ACE2-overexpressed 293T cells. Two-way ANOVA with Tukey's multiple comparisons test was used to assess statistical significance. g-h, T cell response of WT-LNP mRNA vaccinated animals (n = 4). CD8+ (g) and CD4+ (h) T cell responses were measured by intracellular cytokine staining 6 hours after the addition of BFA. The unpaired parametric t-test was used to evaluate the statistical significance.
Immune responses to variant-specific vaccines
Both variant-specific vaccines produced strong antibody responses against wild-type and tested variants, just like the wild-type vaccine. In addition, both variant-specific vaccines induced robust neutralizing antibodies against all tested variants. The binding and neutralizing antibody titers were significantly higher after booster immunization compared to that after prime immunization.
Specifically, the B.1.351-specific vaccine showed similar neutralizing efficacy against all tested viral variants. However, the B.1.617-specific vaccine induced significantly higher antibody titers against the B.1.617 variant compared to that against the wild-type virus and B.1.351 variant.
Regarding cellular immunity, both variant-specific vaccines induced spike-specific CD8+ and CD4+ immune responses similar to the wild-type vaccine.
Immune cell transcriptional landscape following variant-specific vaccination
The findings of single-cell transcriptomics sequencing revealed that both variant-specific vaccines caused a significant increase in activated CD8+ T cells and a slight decrease in natural killer cells. In addition, the B.1.351-specific vaccine caused a slight increase in dendritic cells.
The analysis identified differential expressions of many genes regarding vaccine-induced transcriptomic changes in the B cell and T cell subpopulations. In B cells, differentially expressed genes were associated with B cell activation, immune effector, and immune response. Similarly, in CD4+ and CD8+ T cells, differentially expressed genes were associated with T cell activation, immune effector, and immune response. For both vaccines, the most significantly upregulated genes in B and T cells represented transcription and translation machinery, indicating active lymphocyte activation following vaccination.
B cell and T cell receptor diversity following variant-specific vaccination
The findings of single-cell and bulk B/T cell receptor sequencing revealed a reduction in clonal diversity in the spleen, peripheral blood, and lymph node samples following variant-specific vaccination. These observations indicate that both B.1.351- and B.1.617-specific vaccines cause expansion of a small number of B cell and T cell clones.
Study significance
The study highlights that variant-specific COVID-19 vaccines induce robust humoral and cellular immune responses and are associated with a strong signature of transcriptional and translational machineries in lymphocytes.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Peng L. 2021. Systems immune profiling of variant-specific vaccination against SARS-CoV-2, bioRxiv, https://www.biorxiv.org/content/10.1101/2021.12.02.471028v1
- Peer reviewed and published scientific report.
Peng, Lei, Paul A. Renauer, Arya Ökten, Zhenhao Fang, Jonathan J. Park, Xiaoyu Zhou, Qianqian Lin, et al. 2022. “Variant-Specific Vaccination Induces Systems Immune Responses and Potent in Vivo Protection against SARS-CoV-2.” Cell Reports Medicine 3 (5): 100634. https://doi.org/10.1016/j.xcrm.2022.100634. https://www.cell.com/cell-reports-medicine/fulltext/S2666-3791(22)00159-8.