Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), since its emergence, has continuously evolved to evade host hostilities as well as increase transmission by raising new variants of concerns (VOCs). Virus host adaptation is evident by the rise of VOCs (beta, beta, gamma, and delta variants) that impede the neutralization effect of antibodies. Recently (Nov. 24th, 2021), a novel strain of SARS-CoV-2 named Omicron emerged in South Africa and quickly spread worldwide.
Currently, scientists are trying to understand how Omicron is spread, and whether current therapeutics will still be effective against it. Researchers studied the binding strength of Omicron with ACE2 and seven monoclonal antibodies either approved by the FDA or undergoing phase III clinical trials, in a study published on the bioRxiv* preprint server.
 Study: Omicron: A heavily mutated SARS-CoV-2 variant exhibits stronger binding to ACE2 and potently escape approved COVID-19 therapeutic antibodies. Image Credit: NIAID
Study: Omicron: A heavily mutated SARS-CoV-2 variant exhibits stronger binding to ACE2 and potently escape approved COVID-19 therapeutic antibodies. Image Credit: NIAID

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The Omicron Variant
The Omicron variant has many novel mutations in both structural and non-structural proteins. For example, scientists have observed more than 32 mutations in the Spike protein alone, with 15 of these residing in the receptor-binding domain (RBD). Such a large number of mutations have raised concerns over increased transmissibility, immune escape, and vaccine failure.
The non-structural proteins encoded by the ORF1ab contain mutations in the nsp3, nsp4, nsp5, nsp6, nsp12, and nsp14. In addition, Omicron harbors mutations in the other structural proteins, including Envelope (E), Membrane (M), and Nucleocapsid (N). Since N is highly immunogenic, these mutations could help escape the host immune response. About half of the mutations possess the potential to dampen the potency of therapeutic and enhance ACE2 binding. A significant cause for concern is that this variant can infect vaccinated people, as has been demonstrated by vaccinated people in South Africa and Hong Kong being affected.
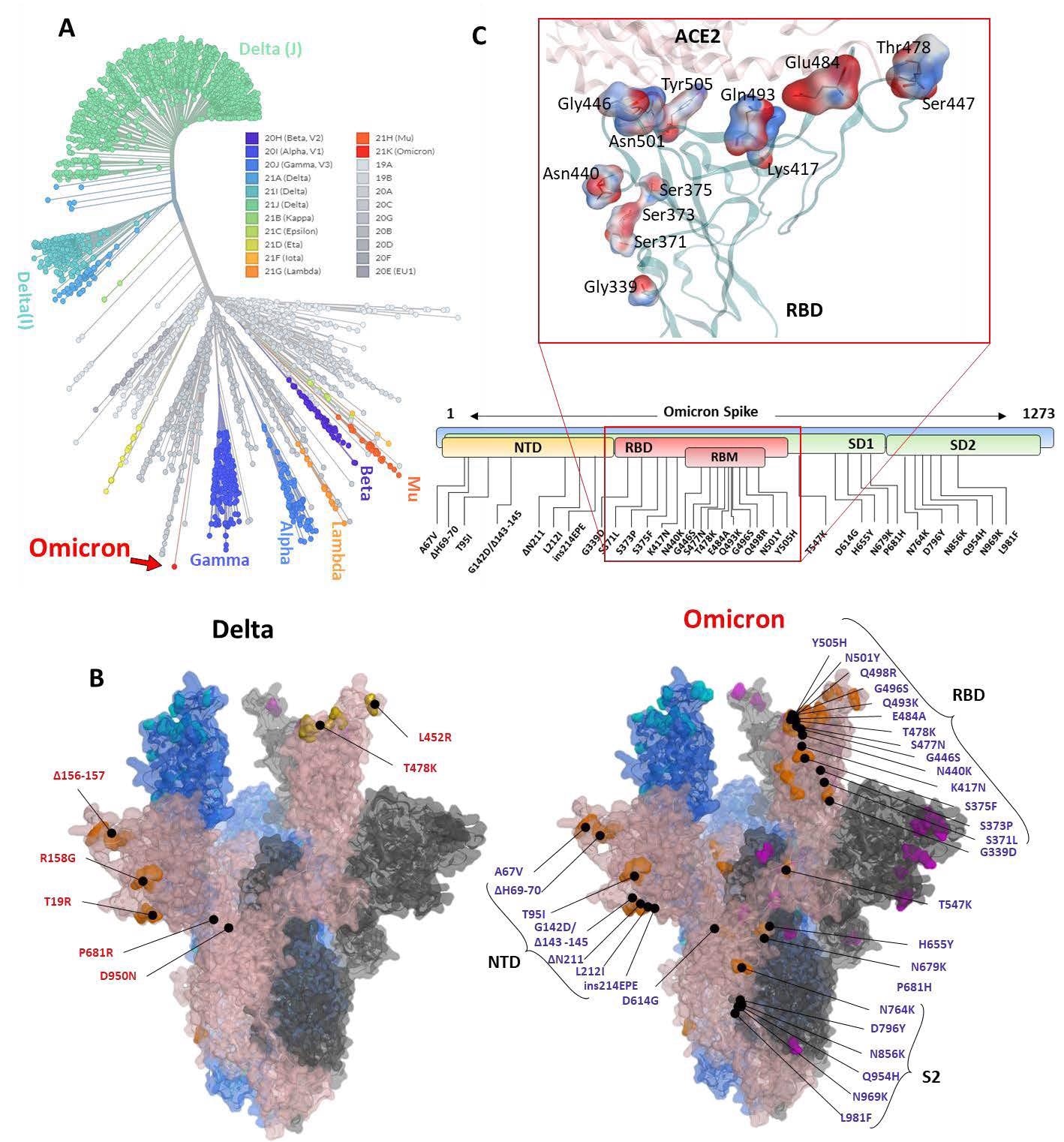 Phylogeny of the Omicron and annotation of the mutation in Spike protein. The Unrooted phylogenetic tree was constructed from the Nextstrain servers. Wuhan-Hu-1/2019 strains were taken as a reference sequence. B) The full-length Delta and Omicron Spike were built to annotate the relative (not exact) positions of the mutations on the surface map of Spike. C) The amino acids mutated in the RBD of Omicron are shown concerning the ACE2 interface. Residues are colored according to the electrostatic map of the WT strain. Respective Omicron mutations are depicted in the panel below the RBD surface map.
Phylogeny of the Omicron and annotation of the mutation in Spike protein. The Unrooted phylogenetic tree was constructed from the Nextstrain servers. Wuhan-Hu-1/2019 strains were taken as a reference sequence. B) The full-length Delta and Omicron Spike were built to annotate the relative (not exact) positions of the mutations on the surface map of Spike. C) The amino acids mutated in the RBD of Omicron are shown concerning the ACE2 interface. Residues are colored according to the electrostatic map of the WT strain. Respective Omicron mutations are depicted in the panel below the RBD surface map.
A New Study
By conducting molecular modeling and mutational analyses, scientists sought to understand how the Omicron variant enhanced its transmissibility and whether it can escape the FDA-approved Spike-neutralizing COVID-19 therapeutic antibodies.
The researchers selected seven therapeutic antibodies, including Etesevimab, Bamlanivimab, AZD8895, AZD1061, Imdevimab, Casirivimab, and CT-p59.
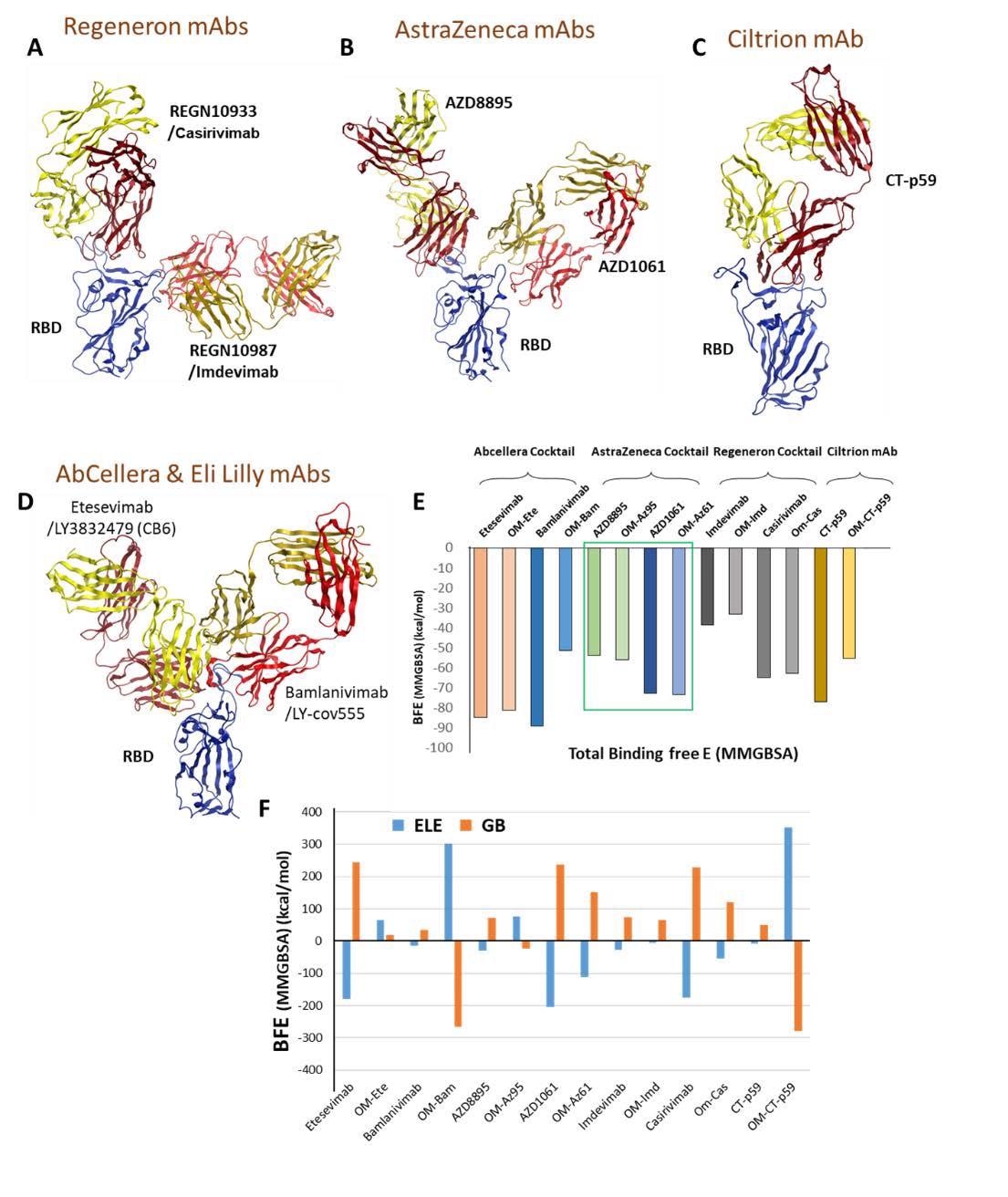 Mutations in the Omicron RBD distort the epitopes of therapeutic mAbs. A-D) Crude epitopes of seven selected mAbs are shown on the RBD. Antibodies used as cocktails are labeled with their sponsors. All variable light chains are colored yellow or orange and variable heavy chains are colored red. E) Changes in the binding affinity of the RBDOmic-mAbs relative to RBDWT-mAbs are shown. The Binding energies were calculated through endpoint MM/GBSA). F) Changes in the electrostatic potentials and polar solvation energies are shown to each RBD-mAb complex.
Mutations in the Omicron RBD distort the epitopes of therapeutic mAbs. A-D) Crude epitopes of seven selected mAbs are shown on the RBD. Antibodies used as cocktails are labeled with their sponsors. All variable light chains are colored yellow or orange and variable heavy chains are colored red. E) Changes in the binding affinity of the RBDOmic-mAbs relative to RBDWT-mAbs are shown. The Binding energies were calculated through endpoint MM/GBSA). F) Changes in the electrostatic potentials and polar solvation energies are shown to each RBD-mAb complex.
Main Findings
The spike-ACE2 interaction: Researchers observed three deletion sites in the N-terminus domains (NTD) and at least 15 substitutions in the RBD region in the Omicron variant. The Spike also harbors mutations, such as K417, T478, E484, and N501, which have been reported in previous VOCs.
An important observation was that at least 11 (out of the 15) mutated residues could influence ACE2 binding and significantly affect the binding affinity.
The Omicron spike (compared to the prototype SARS-CoV-2) had three deletions, i.e., Δ69-70, Δ143-145, and Δ211, and one highly charged insertion at 214 positions in the Spike, i.e., ins214EPE.
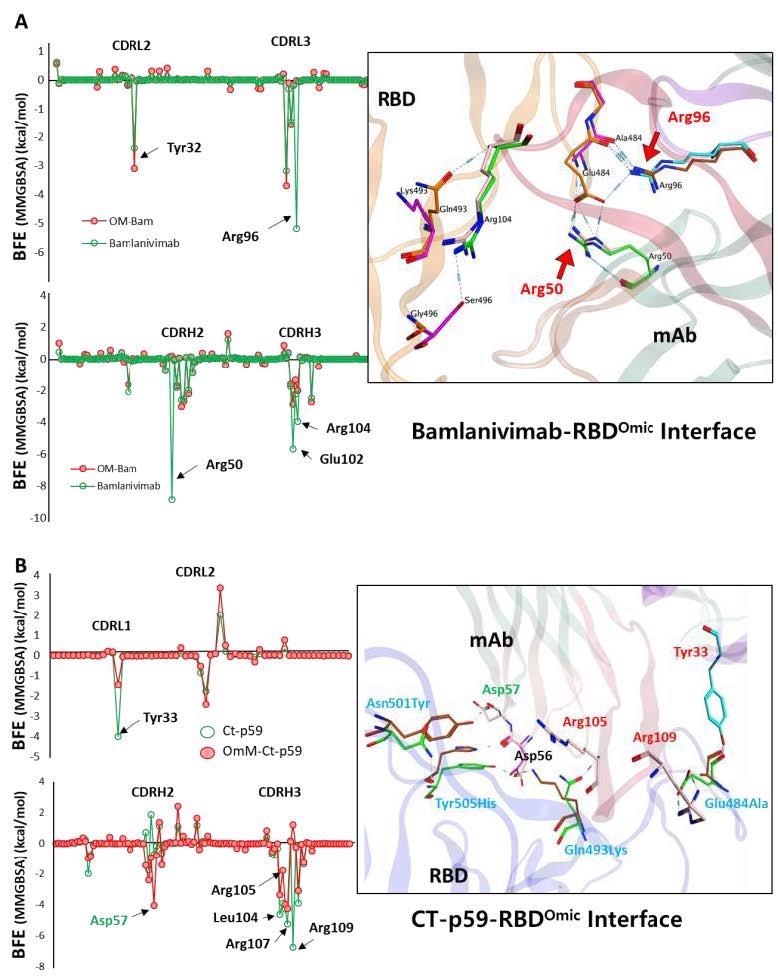 Per-residue changes in the binding affinity of RBD-mAbs were monitored, and the hotspots on CDRs of (A) Bamlanivimab and (B) CT-p59 are labeled. The change in the hydrogen bonds network of the selected hotspots is shown at the right.
Per-residue changes in the binding affinity of RBD-mAbs were monitored, and the hotspots on CDRs of (A) Bamlanivimab and (B) CT-p59 are labeled. The change in the hydrogen bonds network of the selected hotspots is shown at the right.
In order to monitor the relative binding strength of RBD-ACE2 complexes of both the prototype Wuhan and Omicron strains, scientists used a protein design strategy and calculated binding affinity and stability changes.
Individually substituted residues were seen to have a slight effect on the local stability of the RBD-ACE2 complexes. However, a large increase in the binding affinity by T478K, Q493K, and Q498R led to an overall increase in the binding affinity of the RBDOmic with ACE2.
Researchers studied the change in electrostatic potential of the RBDOmic relative to that of RBDWT. The electrostatic energy of ACE2-RBDOmic was found to be double that of ACE2-RBDWT. The energy distribution suggested that mutations in the RBDOmic directly enhance the binding strength of amino acids in the same network. Overall, the data showed that Omicron binds to ACE2 with greater affinity, partly explaining its increased transmissibility. However, the pathogenicity of the new strain could not be predicted.
Binding of therapeutic antibodies: Scientists constructed structural models of seven mAbs bound to RBDOmic and showed a substantial drop in the total binding energies of Bamlanivimab and CT-p59. Except for AZD1061 (AstraZeneca), all other mAbs showed a significant drop.
Changes in energies were calculated for CT-p59 and Bamlanivimab to gain a better understanding of the mutations involved in weakening the RBDOmic -mAbs interactions.
It was observed that R96 and R50 of the Bamlanivimab, which establish highly stable salt bridges with the E484 of RBDWT, completely lost their binding upon E484A mutation. E102 and R104 in CDRH3 also showed a 50% reduction in binding energies. E484A, Q493K, and Y505H mutations in RBDOmic were responsible for the lowering of the bindings.
Overall, these data raise grave concerns about the efficacy of therapeutic mAbs in Omicron patients.
Concluding Remarks
The Omicron variant has been proven to be more transmissible than the Delta variation, and the threat is now global. Therefore, there is an urgent need to closely monitor the COVID-19 variant and accelerate vaccination, as this could reduce COVID-19 infections dramatically. Additionally, the search for more effective therapeutics must continue.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources