Variants of concern, including the dominant Delta variant, possess mutations that may change the sequence diversity during diagnostic testing — leading to false-negative results. The findings may help prevent infected individuals with false-negative results from coming into contact with others and further transmitting the virus.
 Study: RT-PCR/MALDI-TOF diagnostic target performance reflects circulating SARS-CoV-2 variant diversity in New York City. Image Credit: NIAID
Study: RT-PCR/MALDI-TOF diagnostic target performance reflects circulating SARS-CoV-2 variant diversity in New York City. Image Credit: NIAID

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
How they did it
The research team collected 448 specimens that were positive for SARS-CoV-2 from April 11, 2021, to August 28, 2021, from Mount Sinai. They compared the PCR diagnostic test results with data from 256 genomes collected from the specimens.
The genomes from SARS-CoV-2-positive samples were used to find primer/probe binding sites (PBS) mismatches in the nucleocapsid gene as well as any sequence diversity that could impact diagnostic performance for the Agena MassARRAY SARS-CoV-2 Panel.
The testing involved identifying a maximum of five viral targets, including three targets in the nucleocapsid gene (N1, N2, and N3) as well as two in the ORF1ab gene (ORF1A and ORF1AB). Diagnostics will display a positive SARS-CoV-2 result if two or more targets are detected. Conversely, results are negative if less than two targets are detected.
Diagnostic results and detection rates
Approximately 26% of SARS-CoV-2-positive specimens had missed one or more viral targets. Sequencing the available 256 SARS-CoV-2 positive samples detected all five viral targets in 38% of samples. The others had diagnostic misses of one or more targets.
The researchers also looked at weekly detection rates of viral targets that would yield a positive diagnostic result for SARS-CoV-2.
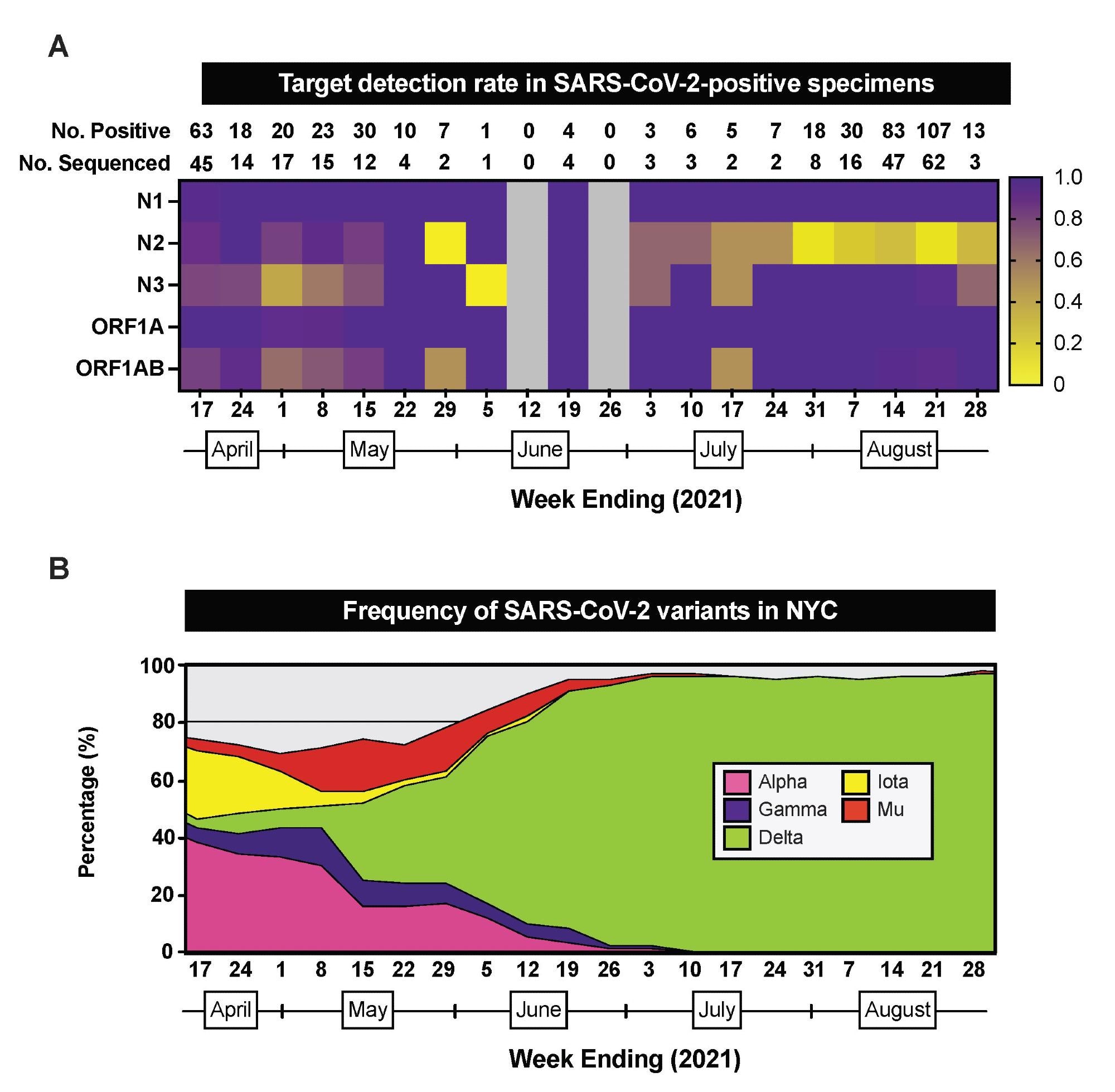 RT-PCR/MALDI-TOF target detection rate in SARS-CoV-2-positive specimens. (A) Heatmap depicting the proportion of SARS-CoV-2-positive specimens that have detectable RT PCR/MALDI-TOF targets (N1, N2, N3, ORF1A, ORF1AB) by week from April 11 through August 28, 2021. The total number of SARS-CoV-2-positive specimens and the number of SARS-CoV-2-positive specimens sequenced by pathogen surveillance are depicted above each week (column). Grey boxes indicate weeks where no specimens were positive for SARS-CoV-2 on this platform. (B) Stacked area plots depicting frequencies of SARS-CoV-2 variants reported by publicly-available NYC Department of Health surveillance data within the same timeframe.
RT-PCR/MALDI-TOF target detection rate in SARS-CoV-2-positive specimens. (A) Heatmap depicting the proportion of SARS-CoV-2-positive specimens that have detectable RT PCR/MALDI-TOF targets (N1, N2, N3, ORF1A, ORF1AB) by week from April 11 through August 28, 2021. The total number of SARS-CoV-2-positive specimens and the number of SARS-CoV-2-positive specimens sequenced by pathogen surveillance are depicted above each week (column). Grey boxes indicate weeks where no specimens were positive for SARS-CoV-2 on this platform. (B) Stacked area plots depicting frequencies of SARS-CoV-2 variants reported by publicly-available NYC Department of Health surveillance data within the same timeframe.
Weekly testing shows the lowest detection rates for the N2 and N3 targets on the nucleocapsid gene.
Sequencing data also show the lowest detection rate for N2 during the week of July 31 and the lowest detection rate for N3 during the week of May 1.
The changes in detection rates for both targets suggest the presence of different SARS-CoV-2 variants in the area.
In New York City, the N3 detection rate was lowest when Alpha made up 33% of COVID-19 cases. Conversely, when 96% of circulating SARS-CoV-2 variants were Delta, N2 had the lowest detection rate.
Mismatches on SARS-CoV-2 genome sequences impact diagnostic performance
Next, the researchers used diagnostic targeting data and genome sequencing to find mismatches for each detected and undetected target. Doing so would shed light on how SARS-CoV-2 genomic diversity alters diagnostic results.
They found that the presence of mismatches could affect primer/probes binding to target sites.
When there were zero mismatches between genome sequences, diagnostic testing found four targets — N1, N3, ORF1A, and ORF1AB.
With the exception of the N1 reverse, N3 reverse, and the ORF1AB primers with zero mismatches, there was at least one mismatch for every target.
When the N2 target was identified, 1 to 3 mismatches were also detected. Samples that could not detect N2 had a significantly higher number of mismatches. A similar pattern was observed with N3.
The researchers then calculated the nucleotide diversity per site for each sequence to gain insight into SARS-CoV-2 sequence diversity.
A seven-fold increase in diversity occurred at the N2 target site, followed by the N3 target. The genomic diversity paralleled the increasing presence of Delta (56.7%), Alpha (15.7%), and the Iota (16.5%) variant.
There were also individual mismatches linked with undetected targets. For instance, mismatches on the first to third base pair of the N2 forward primer PBS in genomes led to variability in detection.
About 192 specimens had at least one mismatch to the third base pair in the N2 forward primer. Approximately 26% had three substitutions in the nucleocapsid gene, and the remaining 74% had a G28881U substitution.
In 123 genomes with undetected N2 targets, about 90% had a mismatch at the terminal 3’ base of the N2 probe. Additionally, in 36 samples with undetected N3 targets, about 69% showed a mismatch on the 3’ end of the N3 primer.
Target dropout based on lineage of SARS-CoV-2 variant
The samples were collected at a time when the Alpha variant was prevalent in New York City. The researchers analyzed the genomes of specimens paying close attention to those with a G28916U substitution.
The G28916U polymorphism creates a G215C amino acid substitution in the nucleocapsid protein.
In a global dataset of genomes, they found 62% of all genomes related to the Delta variant. About 82% of these genomes had the G28916U substitution.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Hernandez MM, et al. (2021). RT-PCR/MALDI-TOF diagnostic target performance reflects circulating SARS-CoV-2 variant diversity in New York City. medRxiv. Doi: https://doi.org/10.1101/2021.12.04.21267265, https://www.medrxiv.org/content/10.1101/2021.12.04.21267265v1
- Peer reviewed and published scientific report.
Hernandez, Matthew M., Radhika Banu, Ana S. Gonzalez-Reiche, Brandon Gray, Paras Shrestha, Liyong Cao, Feng Chen, et al. 2022. “RT-PCR/MALDI-TOF Diagnostic Target Performance Reflects Circulating SARS-CoV-2 Variant Diversity in New York City.” The Journal of Molecular Diagnostics 24 (7): 738–49. https://doi.org/10.1016/j.jmoldx.2022.04.003. https://www.jmdjournal.org/article/S1525-1578(22)00108-8.