Firstly, I felt that neuroscience is one of the areas of biology that is still mostly unsolved and there’s lots to do. Secondly, there was a huge technological aspect to it: in neuroscience, there are a lot of techniques involved and analysis of large amounts of data, which fit well with my background.
TM: My name is Troy Margrie and I am a Professor of Systems Neuroscience at UCL and have been involved in experimental neuroscience for more than 25 years. From a very young age, I have been interested in how we and other animals learn. It all started with me spending a lot of time playing with my pet dog and teaching it various tricks.
I started thinking about how things such as the environment and reinforcement influence his behavior. That was the starting point, then like many other scientists, I had a very inspiring high school biology teacher who taught me to think about nature in a Darwinistic way and how such principles might even shape an individual organisms’ behaviors. Those two things are probably the key factors that got me interested in what the brain can do and how it informs us about the world around us.
Image Credit: MattLphotography/Shutterstock.com
How has the field of neuroscience changed over the last 20 years? What role has new technology played in this?
TM: I would say genetics has probably had the most significant impact. From flies to fish and mammals, it has enabled us to nail down particular cell types and circuits in complex systems that are otherwise very difficult to dismantle and understand their individual elements.
Now we can even use genetics to activate and silence specific circuits in the brain while the animal is performing a particular task. Without these tools, we would still be recording in the dark with limited ability to functionally target specific neurons or circuits. More recently other types of technologies, such as machine learning-based algorithms help us analyze and understand data, but I think the application of genetic tools has had the most impact on my generation.
BrainGlobe is an open-source platform for computational neuroanatomy. Please could you tell us more about the BrainGlobe platform and how it works?
AT: All brains are different, but we still need to study many different brains to understand a particular phenomenon. BrainGlobe provides tools to allow you to both extract information from images of brains and also to pull them together into a common average to build up atlases of brain structure and function.
We provide tools to allow researchers to develop their own 3D models of how the brain is working and we also provide tools that stretch across different fields of neuroscience, allowing researchers studying different animals to bring their results together.
What are some of the tools available to use at BrainGlobe? Are there any initiatives/applications that have especially benefitted from BrainGlobe?
AT: BrainGlobe includes a tool called cellfinder which allows you to identify and locate individual cells marked by genetics in large images of entire brains. There’s another tool called brainrender which allows you to visualize data from multiple brains together to explore in a single model of the brain.
The best example of an initiative that I think especially benefitted from BrainGlobe is described in a spinal connectome paper (Wang et al. 2021), where the entire paper is based on results that used our analysis and visualization tools.
3D rendering of labelled mouse brain cells detected by cellfinder
As an open-source platform, researchers around the world can use and adapt your platform for their own research. What are the advantages of this for not only researchers but scientific advancements as a whole?
TM: In the past, we literally had a physical book or a PDF containing images and drawings of the mouse brain. You would have to flick through this book until you found what you estimate to be the correct page (i.e. brain section) in the book that matched the piece of tissue that you had in front of you and then you’d go about trying to assign where, in this image on this page in the book, your cells were located. This was a very subjective process of attributing a brain area to the cells you were interested in.
Now with these high-resolution digital atlases and the tools that we and others at SWC have developed, everyone can locate their cells of interest within the same atlas space. That means I can directly compare where my cell falls in the atlas with where another researcher’s cells are located within the very same atlas, without either of us having to go through this subjective process of attributing a brain circuit to the cells of interest. This means we can now more directly compare results with one another, whereas previously that wasn’t very straightforward.
AT: The fact that the tools are open-source themselves means that they are free so more researchers can use them, but also anybody can contribute to the software. This means the software is not limited by the applications at the SWC or the problems we face, but anyone can add new features so the software can grow and be of greater use to the community.
You have recently been awarded two grants from the Chan Zuckerberg Initiative (CZI). How will BrainGlobe invest this support, and how important is funding to helping the progression of innovative technologies?
AT: We are using this grant to make these tools as easy to use as possible. The funding is to develop napari plugins – napari is an image viewer that’s used in a few areas of science now. What we will be building are easy-to-use, flexible, graphical user-interfaces for the software so that researchers, regardless of the amount of programming experience that they have, can apply these tools to their own research and their own data.
Traditionally funding in the life sciences is based around a biological question, which is important, but there’s a huge amount of technological research that needs to go into answering these questions and developing new tools. Tools often come out of labs like Troy’s, but then there’s limited funding to really maintain the software and build them for use in the community, rather than just a single lab. And so this kind of dedicated funding is very important.
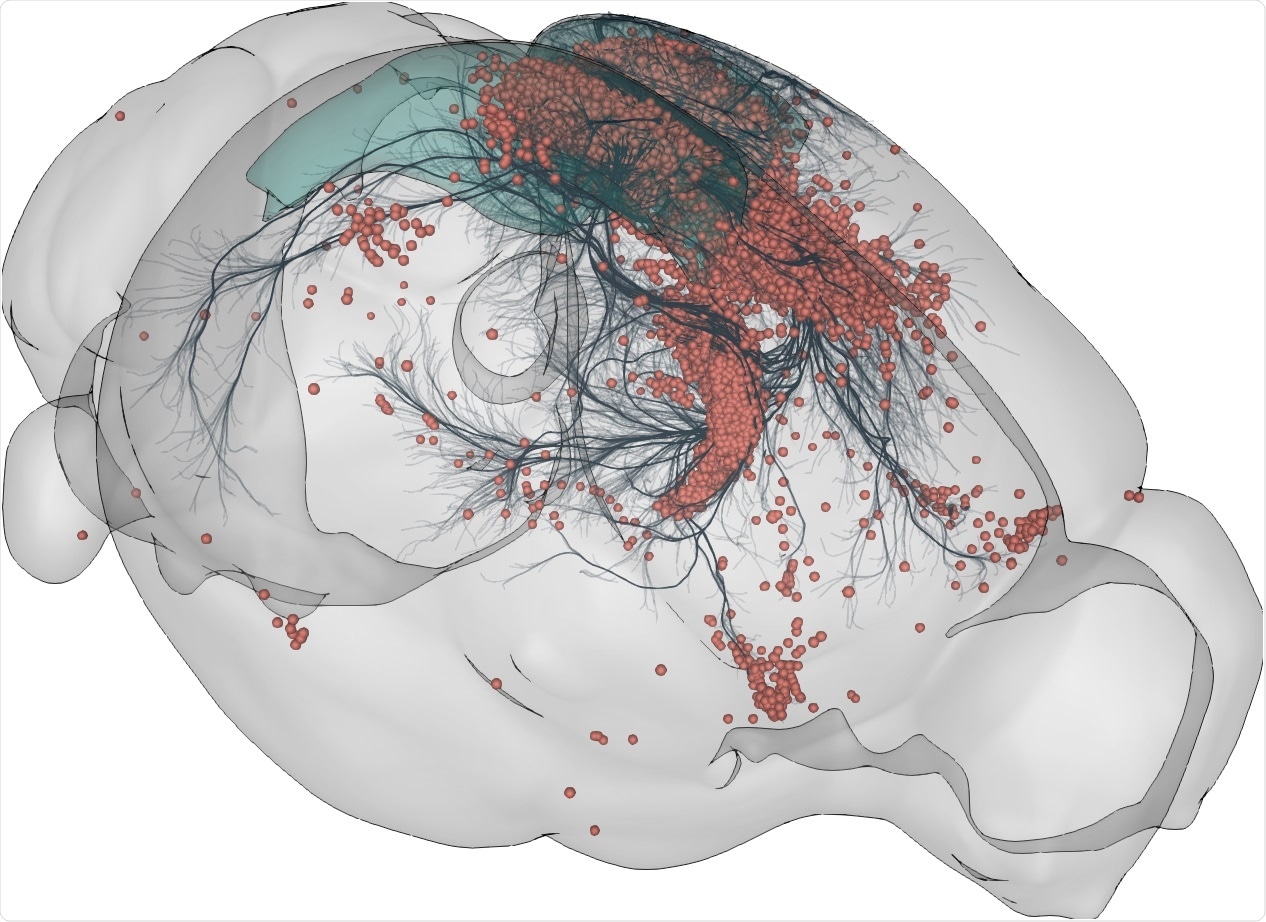
Image Credit: Sainsbury Wellcome Centre
What has been the most exciting project you have been involved with at BrainGlobe?
TM: For me, the most exciting part was the experiments that Christian Niedworok did at the National Institute for Medical Research. He was setting up a pipeline to automatically register a 3D image set to a digital atlas. The most interesting part was that we showed the algorithm was no worse, and perfectly reliable, compared to human beings.
We asked lots of scientists to find the right page in the book that corresponds to the slide image we gave them. We compared across human beings, but we also tricked individuals and gave the same image twice in some cases.
We showed that not only was there a significant amount of variability between individuals, but when you ask the same individual to repeat the task, it is just as variable as asking two people to do the task. So there’s a reliability issue there and there’s a variability issue there across the scientists. This nicely highlights the scientific challenges that we faced and how these new computational neuroanatomy tools can address this problem.
AT: The most interesting aspect for me is the sociology of the BrainGlobe project. Not just in terms of seeing how people are applying the software but also seeing how people adopt these tools and build their own tools based on ours.
It is fascinating to see how people adapt the tools and add new features. For me, the community element and seeing how people engage with the project has been the most interesting.
The ongoing COVID-19 pandemic has shown us that medical and scientific advancements can be made quickly when everyone works together and shares science. How can we take this message and encourage more researchers and organizations to work collaboratively?
TM: The challenging part is getting people to recognize that there’s mutual benefit to working together. I think the message that we could consider trying to propagate to the community is that by building tools that work within a coherent framework, we can achieve analytical approaches that analyze datasets in a way that we can all agree are valid. So not only have the tools been validated, but the way we express ourselves as scientists and talk about this data is a valid approach because we’re all singing from the same sheet.
The problem is that people need to be incentivized to work together so you need to think about what incentivizes people before you just send out a message to say collaborating is best for all of us. In this case, what people really get out of it is a free tool that works really well and helps them to interrogate and disseminate their data in a way that others can digest and believe, which historically in neuroanatomy has been very challenging. When you take away a lot of the subjectivity from the analysis, the data should become more rigorous and that is good for everyone and is in the interest of the science.
AT: We can incentivize people by working with them to adapt the software to help them with their experimental questions. The community aspect also allows people to bring their data together in the same framework, which shares it in a way that’s more easily understandable. For example, we’re all used to understanding a map of the UK. If we have a common map of the brain, a common atlas, then it is easier to understand data if it is in that model.
This also means that people can access data from other places as well. For example, a researcher might have been studying one brain area and they want to compare it to another brain area. Perhaps another lab has already done those experiments in the other area and shared their data in this common model, so it is easier to compare results and people don’t have to repeat experiments unnecessarily.
Have you got any exciting projects upcoming at BrainGlobe? If so, what are they?
AT: We have a few technical projects and new software in the pipeline but I think there are two main projects that I am most excited about. Firstly, the work that we will be doing under the grant is to make the software as easy to use as possible and also to try and engage with other research communities that might not have used the software yet.
The second part is adapting the software so it can be used in more areas of research. There are different models for different animal species that researchers are studying and the more of these atlases that we can bring into the software, the more useful this software can be to a wider audience.
TM: What’s also interesting is that it is also starting to draw the attention of people outside of neuroscience. One of the tools in BrainGlobe is a cell detection algorithm and there’s a brain segment algorithm that allows you to identify objects within brain space.
There’s interest evolving from cancer research groups who want to be able to identify tumors and find the location of tumors and their size and shape. So I think there are applications beyond the brain that BrainGlobe in principle could deliver on and that’s very exciting.

Image Credit: metamorworks/Shutterstock.com
Artificial intelligence and machine learning platforms have become increasingly involved in a range of sectors. Knowing the impact they could have in the biomedical sector, what do you think the future of neuroscience will look like as technology develops?
AT: I don’t think it is going to be as immediately transformative as people think because AI at the moment is still not that intelligent, but I hope that it will essentially free up time for researchers. There’s still a lot of research that is very manual and takes painstaking effort to go through data and I hope that increasingly more of this can be automated to allow researchers to spend more time doing experiments and thinking and asking better questions rather than spending hours at their computer analyzing data.
TM: One limitation is that we still don’t know how machine learning works on many datasets – we often don’t know what aspects of the data that these algorithms are using to refine the analysis of the data. Despite this, it is very clear that AI can do things very well that would normally take us a lot of time or may not even be possible to do manually.
For example, behavioral analysis involves quantifying what an animal is doing as it is living in an arena, and we can now use machine learning to track very specific aspects of the animal’s posture and behavior. Previously this was a very subjective approach.
Now, once you have your training dataset, you can run your videos through the tool and you can spend your time doing something else rather than having this burden of many hundreds and thousands of hours of video analysis. In some ways, AI and machine learning are giving us more time to think about the brain versus having to spend a lot of your time analyzing data that you’ve collected from the brain.
Where can readers find more information?
For more information about BrainGlobe and the Chan Zuckerberg awards, please visit:
About Professor Troy Margrie
Troy’s research is focused on understanding how the activity of cells and circuits contribute to sensory representation, decision-making, and behavior. The lab utilizes a variety of in vivo and in vitro methods including electrophysiology and imaging in behaving mice, which, when combined with viral tracers, can be used to identify both local and long-range connectivity profiles of target populations.
At present, the lab’s primary interest focuses on the role of the retrosplenial cortex, given its widespread projection profile into primary sensory areas such as the visual cortex. In addition, the lab is interested in how sensory experience and life history alter the properties of cortical cells and the extent to which this impacts their sensory function.
Troy is also Associate Director of the SWC and enjoyed the challenge of building the Centre from its beginning to ensure the best possible facilities were provided. He works with the Director, Chief Scientific Officer, and Centre Manager to manage operations across the building and support the science carried out.
About Dr. Adam Tyson
Adam was previously a neuroscientist and software developer in the Margrie lab and is now Scientific Software Lead at the Institute of Cancer Research. Following a Ph.D. in neuroscience at King’s College London and a brief stint at the Institute of Cancer Research, he moved to UCL to develop open-source neuroscience data analysis tools.
Adam co-developed the cellfinder package for the analysis of whole-brain microscopy data and co-founded the BrainGlobe initiative to develop tools and open standards for computational neuroanatomy.